3K84
 
 | | Crystal Structure Analysis of a Oleyl/Oxadiazole/pyridine Inhibitor Bound to a Humanized Variant of Fatty Acid Amide Hydrolase | | Descriptor: | (9Z)-1-(5-pyridin-2-yl-1,3,4-oxadiazol-2-yl)octadec-9-en-1-one, CHLORIDE ION, Fatty-acid amide hydrolase 1 | | Authors: | Mileni, M, Stevens, R.C, Boger, D.L. | | Deposit date: | 2009-10-13 | | Release date: | 2009-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | X-ray crystallographic analysis of alpha-ketoheterocycle inhibitors bound to a humanized variant of fatty acid amide hydrolase.
J.Med.Chem., 53, 2010
|
|
8DW2
 
 | | Cryo-EM structure of SARS-CoV-2 RBD in complex with anti-SARS-CoV-2 DARPin,SR22, and two antibody Fabs, S309 and CR3022 | | Descriptor: | Antibody CR3022 heavy chain, Antibody CR3022 light chain, Antibody S309 heavy chain, ... | | Authors: | Kwon, Y.D, Gorman, J, Kwong, P.D. | | Deposit date: | 2022-07-30 | | Release date: | 2022-12-07 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (4.11 Å) | | Cite: | A potent and broad neutralization of SARS-CoV-2 variants of concern by DARPins.
Nat.Chem.Biol., 19, 2023
|
|
8DW3
 
 | | Cryo-EM structure of SARS-CoV-2 RBD in complex with anti-SARS-CoV-2 DARPin,SR16m, and two antibody Fabs, S309 and CR3022 | | Descriptor: | Anti-SARS-CoV-2 DARPin SR16m, Antibody S309 light chain, Spike protein S1, ... | | Authors: | Kwon, Y.D, Gorman, J, Kwong, P.D. | | Deposit date: | 2022-07-30 | | Release date: | 2022-12-07 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (4.26 Å) | | Cite: | A potent and broad neutralization of SARS-CoV-2 variants of concern by DARPins.
Nat.Chem.Biol., 19, 2023
|
|
3K83
 
 | | Crystal Structure Analysis of a Biphenyl/Oxazole/Carboxypyridine alpha-ketoheterocycle Inhibitor Bound to a Humanized Variant of Fatty Acid Amide Hydrolase | | Descriptor: | 1-DODECANOL, 6-[2-(3-biphenyl-4-ylpropanoyl)-1,3-oxazol-5-yl]pyridine-2-carboxylic acid, CHLORIDE ION, ... | | Authors: | Mileni, M, Stevens, R.C, Boger, D.L. | | Deposit date: | 2009-10-13 | | Release date: | 2009-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | X-ray crystallographic analysis of alpha-ketoheterocycle inhibitors bound to a humanized variant of fatty acid amide hydrolase.
J.Med.Chem., 53, 2010
|
|
6NNG
 
 | | Tubulin-RB3_SLD-TTL in complex with compound DJ95 | | Descriptor: | 2-(1H-indol-6-yl)-4-(3,4,5-trimethoxyphenyl)-1H-imidazo[4,5-c]pyridine, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Kumar, G, Wang, Y, Li, W, White, S.W. | | Deposit date: | 2019-01-15 | | Release date: | 2019-07-10 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Colchicine Binding Site Agent DJ95 Overcomes Drug Resistance and Exhibits Antitumor Efficacy.
Mol.Pharmacol., 96, 2019
|
|
3K39
 
 | |
3J3P
 
 | | Conformational Shift of a Major Poliovirus Antigen Confirmed by Immuno-Cryogenic Electron Microscopy: 135S Poliovirus and C3-Fab Complex | | Descriptor: | C3 antibody, heavy chain, light chain, ... | | Authors: | Lin, J, Cheng, N, Hogle, J.M, Steven, A.C, Belnap, D.M. | | Deposit date: | 2013-04-10 | | Release date: | 2013-07-03 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | Conformational shift of a major poliovirus antigen confirmed by immuno-cryogenic electron microscopy.
J.Immunol., 191, 2013
|
|
8JTS
 
 | | hOCT1 in complex with metformin in outward open conformation | | Descriptor: | Metformin, Solute carrier family 22 member 1 | | Authors: | Zhang, S, Zhu, A, Kong, F, Chen, J, Lan, B, He, G, Gao, K, Cheng, L, Yan, C, Chen, L, Liu, X. | | Deposit date: | 2023-06-22 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Structural insights into human organic cation transporter 1 transport and inhibition.
Cell Discov, 10, 2024
|
|
8JU0
 
 | | hOCT1 in complex with spironolactone in inward facing occluded conformation | | Descriptor: | SPIRONOLACTONE, Solute carrier family 22 member 1 | | Authors: | Zhang, S, Zhu, A, Kong, F, Chen, J, Lan, B, He, G, Gao, K, Cheng, L, Yan, C, Chen, L, Liu, X. | | Deposit date: | 2023-06-23 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural insights into human organic cation transporter 1 transport and inhibition.
Cell Discov, 10, 2024
|
|
8JTW
 
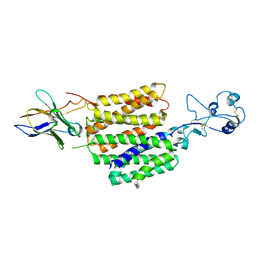 | | hOCT1 in complex with nb5660 in inward facing partially open 1 conformation | | Descriptor: | Solute carrier family 22 member 1, nanobody 56 | | Authors: | Zhang, S, Zhu, A, Kong, F, Chen, J, Lan, B, He, G, Gao, K, Cheng, L, Yan, C, Chen, L, Liu, X. | | Deposit date: | 2023-06-22 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural insights into human organic cation transporter 1 transport and inhibition.
Cell Discov, 10, 2024
|
|
8JTX
 
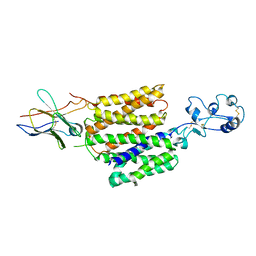 | | hOCT1 in complex with nb5660 in inward facing fully open conformation | | Descriptor: | Solute carrier family 22 member 1, nanobody 56 | | Authors: | Zhang, S, Zhu, A, Kong, F, Chen, J, Lan, B, He, G, Gao, K, Cheng, L, Yan, C, Chen, L, Liu, X. | | Deposit date: | 2023-06-22 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural insights into human organic cation transporter 1 transport and inhibition.
Cell Discov, 10, 2024
|
|
8JTZ
 
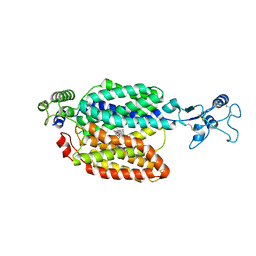 | | hOCT1 in complex with spironolactone in outward facing partially occluded conformation | | Descriptor: | SPIRONOLACTONE, Solute carrier family 22 member 1 | | Authors: | Zhang, S, Zhu, A, Kong, F, Chen, J, Lan, B, He, G, Gao, K, Cheng, L, Yan, C, Chen, L, Liu, X. | | Deposit date: | 2023-06-22 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural insights into human organic cation transporter 1 transport and inhibition.
Cell Discov, 10, 2024
|
|
8K44
 
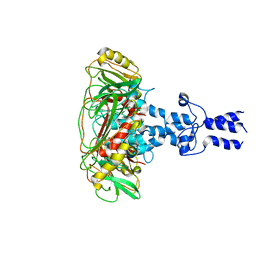 | | Structure of VP9 in Banna virus | | Descriptor: | VP9 | | Authors: | Li, Z, Cao, S. | | Deposit date: | 2023-07-17 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of Banna virus in multiple states reveal stepwise detachment of viral spikes.
Nat Commun, 15, 2024
|
|
8K3S
 
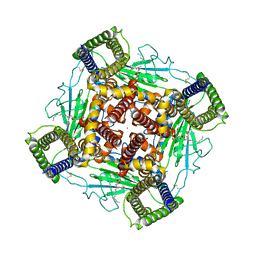 | | Structure of PKD2-F604P complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Chen, M.Y, Su, Q, Wang, Z.F, Yu, Y. | | Deposit date: | 2023-07-16 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular and structural basis of the dual regulation of the polycystin-2 ion channel by small-molecule ligands.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8K4A
 
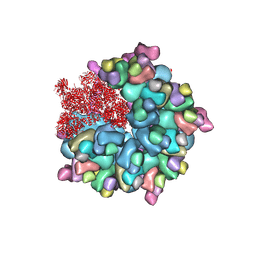 | | Structure of Banna virus core | | Descriptor: | VP10, VP2, VP8 | | Authors: | Li, Z, Cao, S. | | Deposit date: | 2023-07-17 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Cryo-EM structures of Banna virus in multiple states reveal stepwise detachment of viral spikes.
Nat Commun, 15, 2024
|
|
8JTT
 
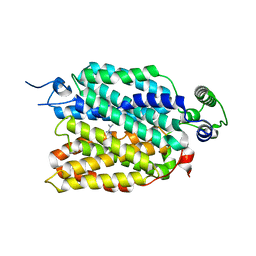 | | hOCT1 in complex with metformin in outward occluded conformation | | Descriptor: | Metformin, Solute carrier family 22 member 1 | | Authors: | Zhang, S, Zhu, A, Kong, F, Chen, J, Lan, B, He, G, Gao, K, Cheng, L, Yan, C, Chen, L, Liu, X. | | Deposit date: | 2023-06-22 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Structural insights into human organic cation transporter 1 transport and inhibition.
Cell Discov, 10, 2024
|
|
8K49
 
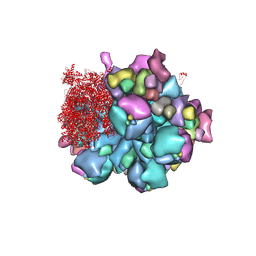 | | Structure of partial Banna virus | | Descriptor: | VP10, VP2, VP4, ... | | Authors: | Li, Z, Cao, S. | | Deposit date: | 2023-07-17 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of Banna virus in multiple states reveal stepwise detachment of viral spikes.
Nat Commun, 15, 2024
|
|
8K43
 
 | |
8JTY
 
 | | hOCT1 in complex with nb5660 in inward facing partially open 2 conformation | | Descriptor: | Solute carrier family 22 member 1, nanobody 56 | | Authors: | Zhang, S, Zhu, A, Kong, F, Chen, J, Lan, B, He, G, Gao, K, Cheng, L, Yan, C, Chen, L, Liu, X. | | Deposit date: | 2023-06-22 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural insights into human organic cation transporter 1 transport and inhibition.
Cell Discov, 10, 2024
|
|
8K42
 
 | | Structure of full Banna virus | | Descriptor: | VP10, VP2, VP4, ... | | Authors: | Li, Z, Cao, S. | | Deposit date: | 2023-07-17 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Cryo-EM structures of Banna virus in multiple states reveal stepwise detachment of viral spikes.
Nat Commun, 15, 2024
|
|
8JTV
 
 | | hOCT1 in complex with metformin in inward occluded conformation | | Descriptor: | Metformin, Solute carrier family 22 member 1 | | Authors: | Zhang, S, Zhu, A, Kong, F, Chen, J, Lan, B, He, G, Gao, K, Cheng, L, Yan, C, Chen, L, Liu, X. | | Deposit date: | 2023-06-22 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural insights into human organic cation transporter 1 transport and inhibition.
Cell Discov, 10, 2024
|
|
3K38
 
 | |
4RFR
 
 | | Complex structure of AlkB/rhein | | Descriptor: | 4,5-dihydroxy-9,10-dioxo-9,10-dihydroanthracene-2-carboxylic acid, Alpha-ketoglutarate-dependent dioxygenase AlkB, MANGANESE (II) ION | | Authors: | Li, Q, Huang, Y, Li, J.F, Yang, C.-G. | | Deposit date: | 2014-09-27 | | Release date: | 2016-04-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Rhein Inhibits AlkB Repair Enzymes and Sensitizes Cells to Methylated DNA Damage.
J.Biol.Chem., 291, 2016
|
|
8E07
 
 | | Crystal structure of HPSE P6 in complex with triose pentosan inhibitor | | Descriptor: | 2,3,4-tri-O-sulfo-beta-D-xylopyranose-(1-4)-2,3-di-O-sulfo-beta-D-xylopyranose-(1-4)-2,3-di-O-sulfo-beta-D-xylopyranose, ACETATE ION, AMMONIUM ION, ... | | Authors: | Whitefield, C, Jackson, C.J. | | Deposit date: | 2022-08-08 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Complex Inhibitory Mechanism of Glycomimetics with Heparanase.
Biochemistry, 62, 2023
|
|
8E08
 
 | | Crystal structure of HPSE P6 in complex with tetraose pentosan inhibitor | | Descriptor: | 2,3,4-tri-O-sulfo-beta-D-xylopyranose-(1-4)-2,3-di-O-sulfo-beta-D-xylopyranose-(1-4)-2,3-di-O-sulfo-beta-D-xylopyranose-(1-4)-2,3-di-O-sulfo-beta-D-xylopyranose, ACETATE ION, Heparanase 50 kDa subunit, ... | | Authors: | Whitefield, C, Jackson, C.J. | | Deposit date: | 2022-08-08 | | Release date: | 2023-07-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Complex Inhibitory Mechanism of Glycomimetics with Heparanase.
Biochemistry, 62, 2023
|
|
