4MN5
 
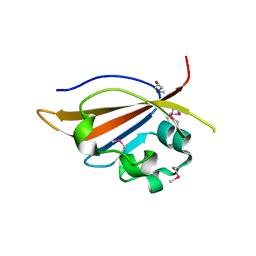 | | Crystal structure of PAS domain of S. aureus YycG | | Descriptor: | Sensor protein kinase WalK, ZINC ION | | Authors: | Shaikh, N, Hvorup, R, Winnen, B, Collins, B.M, King, G.F. | | Deposit date: | 2013-09-10 | | Release date: | 2014-09-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of PAS domain of S. aureus YycG
To be Published
|
|
3FL2
 
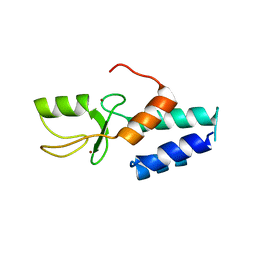 | | Crystal structure of the ring domain of the E3 ubiquitin-protein ligase UHRF1 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-12-18 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the Ring Domain of the E3 Ubiquitin-Protein Ligase Uhrf1
To be Published
|
|
7L7S
 
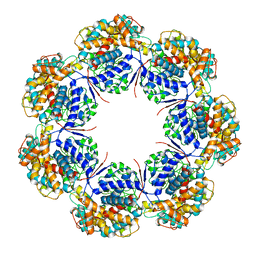 | | Human mitochondrial chaperonin mHsp60 | | Descriptor: | 60 kDa heat shock protein, mitochondrial | | Authors: | Chen, L, Wang, J.C.Y. | | Deposit date: | 2020-12-30 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for the structural dynamics of human mitochondrial chaperonin mHsp60.
Sci Rep, 11, 2021
|
|
6U5H
 
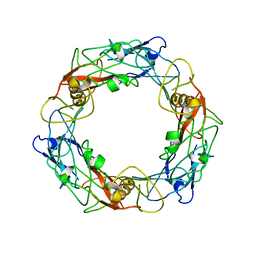 | | CryoEM Structure of Pyocin R2 - precontracted - hub | | Descriptor: | Probable bacteriophage protein Pyocin R2 | | Authors: | Ge, P, Avaylon, J, Scholl, D, Shneider, M.M, Browning, C, Buth, S.A, Plattner, M, Ding, K, Leiman, P.G, Miller, J.F, Zhou, Z.H. | | Deposit date: | 2019-08-27 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Action of a minimal contractile bactericidal nanomachine.
Nature, 580, 2020
|
|
4MML
 
 | | D40A Hfq from Pseudomonas aeruginosa | | Descriptor: | MAGNESIUM ION, Protein hfq, SULFATE ION, ... | | Authors: | Murina, V.N, Filimonov, V.V, Melnik, B.S, Uhlein, M, Mueller, U, Weiss, M, Nikulin, A.D. | | Deposit date: | 2013-09-09 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Effect of conserved intersubunit amino Acid substitutions on hfq protein structure and stability.
Biochemistry Mosc., 79, 2014
|
|
1C1B
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH GCA-186 | | Descriptor: | 6-(3',5'-DIMETHYLBENZYL)-1-ETHOXYMETHYL-5-ISOPROPYLURACIL, HIV-1 REVERSE TRANSCRIPTASE (A-CHAIN), HIV-1 REVERSE TRANSCRIPTASE (B-CHAIN) | | Authors: | Hopkins, A.L, Ren, J, Tanaka, H, Baba, B, Okamato, M, Stuart, D.I, Stammers, D.K. | | Deposit date: | 1999-07-21 | | Release date: | 2000-07-21 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design of MKC-442 (emivirine) analogues with improved activity against drug-resistant HIV mutants.
J.Med.Chem., 42, 1999
|
|
5F8P
 
 | | A Novel Inhibitor of the Obesity-Related Protein FTO | | Descriptor: | 2-OXOGLUTARIC ACID, 4-chloranyl-6-[(2~{S})-6-chloranyl-2,4,4-trimethyl-7-oxidanyl-3~{H}-chromen-2-yl]benzene-1,3-diol, Alpha-ketoglutarate-dependent dioxygenase FTO, ... | | Authors: | Chai, J, Zhou, B, Liu, W, Han, Z. | | Deposit date: | 2015-12-09 | | Release date: | 2016-12-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of FTO-CHTB
To Be Published
|
|
7LFC
 
 | |
7LF4
 
 | |
7LET
 
 | |
7LEQ
 
 | |
5FCW
 
 | | HDAC8 Complexed with a Hydroxamic Acid | | Descriptor: | 4-naphthalen-1-yl-~{N}-oxidanyl-benzamide, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Cole, K.E, Perry, K. | | Deposit date: | 2015-12-15 | | Release date: | 2016-10-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | Structure of 'linkerless' hydroxamic acid inhibitor-HDAC8 complex confirms the formation of an isoform-specific subpocket.
J.Struct.Biol., 195, 2016
|
|
5FPM
 
 | | Structure of heat shock-related 70kDA protein 2 with small-molecule ligand 5-phenyl-1,3,4-oxadiazole-2-thiol (AT809) in an alternate binding site. | | Descriptor: | 5-PHENYL-1,3,4-OXADIAZOLE-2-THIOL, HEAT SHOCK-RELATED 70KDA PROTEIN 2 | | Authors: | Jhoti, H, Ludlow, R.F, Patel, S, Saini, H.K, Tickle, I.J, Verdonk, M. | | Deposit date: | 2015-12-02 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6U5K
 
 | | CryoEM Structure of Pyocin R2 - postcontracted - baseplate | | Descriptor: | Glue PA0627, Sheath Initiator PA0617, Sheath PA0622, ... | | Authors: | Ge, P, Avaylon, J, Scholl, D, Shneider, M.M, Browning, C, Buth, S.A, Plattner, M, Ding, K, Leiman, P.G, Miller, J.F, Zhou, Z.H. | | Deposit date: | 2019-08-27 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Action of a minimal contractile bactericidal nanomachine.
Nature, 580, 2020
|
|
4O4R
 
 | | Murine Norovirus RdRp in complex with PPNDS | | Descriptor: | 3-[(E)-{4-formyl-5-hydroxy-6-methyl-3-[(phosphonooxy)methyl]pyridin-2-yl}diazenyl]-7-nitronaphthalene-1,5-disulfonic acid, RNA-dependent-RNA-polymerase, SULFATE ION | | Authors: | Croci, R, Tarantino, D, Milani, M, Pezzullo, M, Bolognesi, M, Mastrangelo, E. | | Deposit date: | 2013-12-19 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | PPNDS inhibits murine Norovirus RNA-dependent RNA-polymerase mimicking two RNA stacking bases.
Febs Lett., 588, 2014
|
|
5FPT
 
 | | Structure of hepatitis C virus (HCV) full-length NS3 complex with small-molecule ligand 2-(1-methyl-1H-indol-3-yl)acetic acid (AT3437) in an alternate binding site. | | Descriptor: | (1-methyl-1H-indol-3-yl)acetic acid, HEPATITIS C VIRUS FULL-LENGTH NS3 COMPLEX | | Authors: | Jhoti, H, Ludlow, R.F, Saini, H.K, Tickle, I.J, Verdonk, M, Pathuri, P, Williams, P.A. | | Deposit date: | 2015-12-02 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5GG6
 
 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with 8-oxo-dGTP | | Descriptor: | 1,2-ETHANEDIOL, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Arif, S.M, Patil, A.G, Varshney, U, Vijayan, M. | | Deposit date: | 2016-06-15 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Biochemical and structural studies of Mycobacterium smegmatis MutT1, a sanitization enzyme with unusual modes of association
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5GGB
 
 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with 8-oxo-dGDP | | Descriptor: | 2'-deoxy-8-oxoguanosine 5'-(trihydrogen diphosphate), Hydrolase, NUDIX family protein | | Authors: | Arif, S.M, Patil, A.G, Varshney, U, Vijayan, M. | | Deposit date: | 2016-06-15 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Biochemical and structural studies of Mycobacterium smegmatis MutT1, a sanitization enzyme with unusual modes of association
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5GQE
 
 | | Crystal structure of michaelis complex of xylanase mutant (T82A, N127S, and E128H) from Streptomyces olivaceoviridis E-86 | | Descriptor: | Beta-xylanase, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, ... | | Authors: | Suzuki, R, Fujimoto, Z, Kaneko, S, Kuno, A. | | Deposit date: | 2016-08-07 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Azidolysis by the Formation of Stable Ser-His Catalytic Dyad in a Glycoside Hydrolase Family 10 Xylanase Mutant
J.Appl.Glyosci., 65, 2019
|
|
5GL2
 
 | | Crystal structure of TON_0340 in complex with Ca | | Descriptor: | CALCIUM ION, Uncharacterized protein | | Authors: | Lee, S.G, Sohn, Y.S, Oh, B.H. | | Deposit date: | 2016-07-07 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Identification of a Highly Conserved Hypothetical Protein TON_0340 as a Probable Manganese-Dependent Phosphatase.
PLoS ONE, 11, 2016
|
|
4OUF
 
 | | Crystal Structure of CBP bromodomain | | Descriptor: | 1,2-ETHANEDIOL, CREB-binding protein, DI(HYDROXYETHYL)ETHER | | Authors: | Roy, S, Das, C, Tyler, J.K, Kutateladze, T.G. | | Deposit date: | 2014-02-17 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Binding of the histone chaperone ASF1 to the CBP bromodomain promotes histone acetylation.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5G3M
 
 | | Discovery of a novel secreted phospholipase A2 (sPLA2) inhibitor. | | Descriptor: | 4-BENZYLBENZAMIDE, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sandmark, J, Bodin, C, Hallberg, K. | | Deposit date: | 2016-04-29 | | Release date: | 2016-09-14 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of Azd2716: A Novel Secreted Phospholipase A2 (Spla2) Inhibitor for the Treatment of Coronary Artery Disease
Acs Med.Chem.Lett., 7, 2016
|
|
8DGO
 
 | |
1MJB
 
 | | Crystal structure of yeast Esa1 histone acetyltransferase E338Q mutant complexed with acetyl coenzyme A | | Descriptor: | ACETYL COENZYME *A, Esa1 protein | | Authors: | Yan, Y, Harper, S, Speicher, D, Marmorstein, R. | | Deposit date: | 2002-08-27 | | Release date: | 2002-10-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The catalytic mechanism of the ESA1 histone acetyltransferase involves a self-acetylated intermediate.
Nat.Struct.Biol., 9, 2002
|
|
1MJS
 
 | |
