7DY7
 
 | | Discovery of Novel Small-molecule Inhibitors of PD-1/PD-L1 Axis that Promotes PD-L1 Internalization and Degradation | | Descriptor: | 2-[[3-[[5-(2-methyl-3-phenyl-phenyl)-1,3,4-oxadiazol-2-yl]amino]phenyl]methylamino]ethanol, Programmed cell death 1 ligand 1 | | Authors: | Cheng, Y, Wang, T.Y, Lu, M.L, Jiang, S, Xiao, Y.B. | | Deposit date: | 2021-01-20 | | Release date: | 2022-01-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Discovery of Small-Molecule Inhibitors of the PD-1/PD-L1 Axis That Promote PD-L1 Internalization and Degradation.
J.Med.Chem., 65, 2022
|
|
5C3T
 
 | | PD-1 binding domain from human PD-L1 | | Descriptor: | Programmed cell death 1 ligand 1 | | Authors: | Zak, K.M, Dubin, G, Holak, T.A. | | Deposit date: | 2015-06-17 | | Release date: | 2015-11-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Complex of Human Programmed Death 1, PD-1, and Its Ligand PD-L1.
Structure, 23, 2015
|
|
7E9B
 
 | |
5B21
 
 | |
7VYT
 
 | | Crystal structure of human TIGIT(23-129) in complex with the scFv fragment of anti-TIGIT antibody MG1131 | | Descriptor: | CITRATE ANION, MG1131 heavy chain variable region, MG1131 light chain variable region, ... | | Authors: | Jeong, B.-S, Nam, H, Kim, M, Oh, B.-H. | | Deposit date: | 2021-11-15 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural and functional characterization of a monoclonal antibody blocking TIGIT.
Mabs, 14, 2022
|
|
7VUN
 
 | | Design, modification, evaluation and cocrystal studies of novel phthalimides regulating PD-1/PD-L1 interaction | | Descriptor: | (2~{S},3~{S})-2-[[6-[(3-cyanophenyl)methoxy]-2-(2-methyl-3-phenyl-phenyl)-1,3-bis(oxidanylidene)isoindol-5-yl]methylamino]-3-oxidanyl-butanoic acid, Programmed cell death 1 ligand 1 | | Authors: | Cheng, Y, Sun, C.L, Chen, M.R, Yang, P, Xiao, Y.B. | | Deposit date: | 2021-11-03 | | Release date: | 2022-09-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Novel phthalimides regulating PD-1/PD-L1 interaction as potential immunotherapy agents.
Acta Pharm Sin B, 12, 2022
|
|
7VUX
 
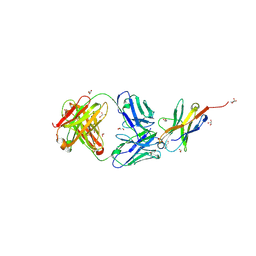 | | Complex structure of PD1 and 609A-Fab | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Huang, H, Zhu, Z, Zhao, J, Jiang, L, Yang, H, Deng, L, Meng, X, Ding, J, Yang, S, Zhao, L, Xu, W, Wang, X. | | Deposit date: | 2021-11-04 | | Release date: | 2021-11-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A strategy for the efficient construction of anti-PD1-based bispecific antibodies with desired IgG-like properties.
Mabs, 14, 2022
|
|
7WVM
 
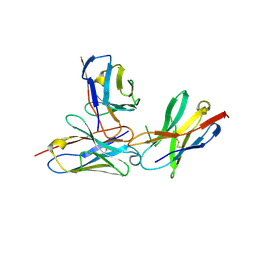 | | The complex structure of PD-1 and cemiplimab | | Descriptor: | Heavy Chain of Cemiplimab, Light Chain of Cemiplimab, Programmed cell death protein 1 | | Authors: | Lu, D, Xu, Z.P, Liu, K.F, Tan, S.G, Gao, G.F, Chai, Y. | | Deposit date: | 2022-02-10 | | Release date: | 2022-04-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | PD-1 N58-Glycosylation-Dependent Binding of Monoclonal Antibody Cemiplimab for Immune Checkpoint Therapy.
Front Immunol, 13, 2022
|
|
7UXO
 
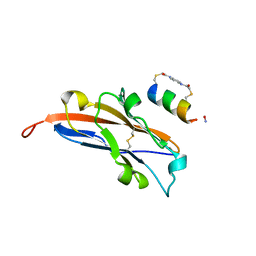 | | Structure of PDL1 in complex with FP30790, a Helicon Polypeptide | | Descriptor: | AMINO GROUP, FP30790, N,N'-(1,4-phenylene)diacetamide, ... | | Authors: | Li, K, Agarwal, S, Tokareva, O, Thomson, T, Travaline, T, Tattersfield, H, Wahl, S, Verdine, G, McGee, J. | | Deposit date: | 2022-05-05 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | De novo mapping of alpha-helix recognition sites on protein surfaces using unbiased libraries.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UX5
 
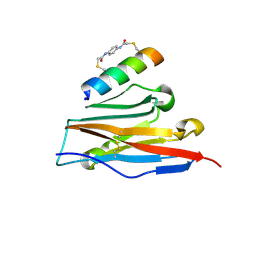 | | Structure of PDL1 in complex with FP28136, a Helicon Polypeptide | | Descriptor: | Helicon FP28136, N,N'-(1,4-phenylene)diacetamide, Programmed cell death 1 ligand 1 | | Authors: | Agarwal, S, Li, K, Tokareva, O, Thomson, T, Travaline, T, Wahl, S, Verdine, G, McGee, J. | | Deposit date: | 2022-05-05 | | Release date: | 2022-12-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | De novo mapping of alpha-helix recognition sites on protein surfaces using unbiased libraries.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UXP
 
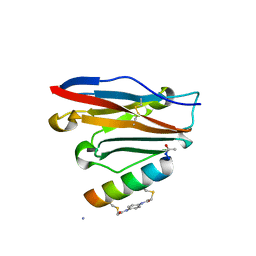 | | Structure of PDL1 in complex with FP28132, a Helicon Polypeptide | | Descriptor: | AMINO GROUP, FP28132, N,N'-(1,4-phenylene)diacetamide, ... | | Authors: | Li, K, Agarwal, S, Tokareva, O, Thomson, T, Travaline, T, Tattersfield, H, Wahl, S, Verdine, G, McGee, J. | | Deposit date: | 2022-05-05 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | De novo mapping of alpha-helix recognition sites on protein surfaces using unbiased libraries.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UXQ
 
 | | Structure of PDL1 in complex with FP28135, a Helicon Polypeptide | | Descriptor: | 1,2-ETHANEDIOL, FP28135, N,N'-(1,4-phenylene)diacetamide, ... | | Authors: | Li, K, Agarwal, S, Tokareva, O, Thomson, T, Travaline, T, Wahl, S, Verdine, G, McGee, J. | | Deposit date: | 2022-05-05 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | De novo mapping of alpha-helix recognition sites on protein surfaces using unbiased libraries.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WSL
 
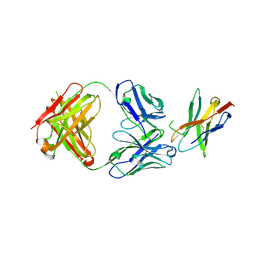 | | PD-1 in complex with Dostarlimab | | Descriptor: | Programmed cell death protein 1, heavy chain, light chain | | Authors: | Heo, Y.S. | | Deposit date: | 2022-01-30 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.534 Å) | | Cite: | Molecular basis of PD-1 blockade by dostarlimab, the FDA-approved antibody for cancer immunotherapy.
Biochem.Biophys.Res.Commun., 599, 2022
|
|
7UVF
 
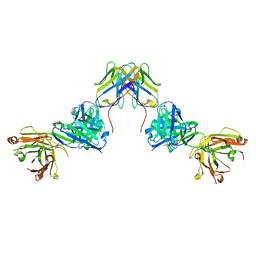 | | Crystal structure of ZED8 Fab complex with CD8 alpha | | Descriptor: | CHLORIDE ION, GLYCEROL, Immunoglobulin heavy chain, ... | | Authors: | Yu, C, Davies, C, Koerber, J.T, Williams, S. | | Deposit date: | 2022-05-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Preclinical development of ZED8, an 89 Zr immuno-PET reagent for monitoring tumor CD8 status in patients undergoing cancer immunotherapy.
Eur J Nucl Med Mol Imaging, 50, 2023
|
|
1A8J
 
 | |
1EAJ
 
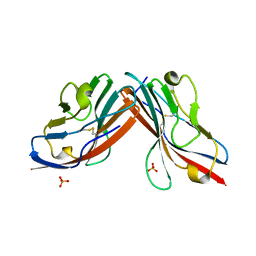 | |
1A6W
 
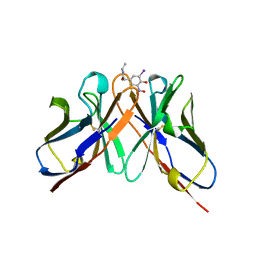 | |
1A7N
 
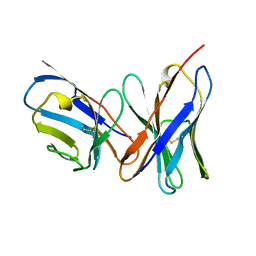 | |
1A7P
 
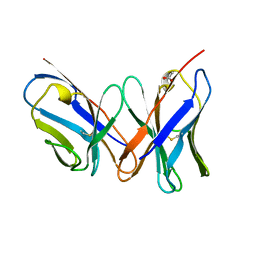 | |
1AC6
 
 | |
1A7Q
 
 | | FV FRAGMENT OF MOUSE MONOCLONAL ANTIBODY D1.3 (BALB/C, IGG1, K) HIGH AFFINITY EXPRESSED VARIANT CONTAINING SER26L->GLY, ILE29L->THR, GLU81L->ASP, THR97L->SER, PRO240H->LEU, ASP258H->ALA, LYS281H->GLU, ASN283H->ASP AND LEU312H->VAL | | Descriptor: | IGG1-KAPPA D1.3 FV (HEAVY CHAIN), IGG1-KAPPA D1.3 FV (LIGHT CHAIN) | | Authors: | Marks, C, Henrick, K, Winter, G. | | Deposit date: | 1998-03-16 | | Release date: | 1998-04-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-Ray Structures of D1.3 Fv Mutants
To be Published
|
|
1AH1
 
 | | CTLA-4, NMR, 20 STRUCTURES | | Descriptor: | CTLA-4, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Metzler, W.J, Bajorath, J, Fenderson, W, Shaw, S.-Y, Peach, R, Constantine, K.L, Naemura, J, Leytze, G, Lavoie, T.B, Mueller, L, Linsley, P.S. | | Deposit date: | 1997-04-11 | | Release date: | 1998-04-15 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human CTLA-4 and delineation of a CD80/CD86 binding site conserved in CD28.
Nat.Struct.Biol., 4, 1997
|
|
1A7O
 
 | |
1A6U
 
 | | B1-8 FV FRAGMENT | | Descriptor: | B1-8 FV (HEAVY CHAIN), B1-8 FV (LIGHT CHAIN) | | Authors: | Simon, T, Henrick, K, Hirshberg, M, Winter, G. | | Deposit date: | 1998-03-03 | | Release date: | 1998-05-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-Ray Structures of Fv Fragment and its (4-Hydroxy-3-Nitrophenyl)Acetate Complex of Murine B1-8 Antibody
To be Published
|
|
1DCL
 
 | |
