5C3D
 
 | | Crystal structure of ABBB + UDP-C-Gal (short soak) + DI | | Descriptor: | (((2S,3R,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl)methyl)phosphonic (((2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl)methyl phosphoric) anhydride, 2-(2-METHOXYETHOXY)ETHANOL, Histo-blood group ABO system transferase, ... | | Authors: | Gagnon, S, Meloncelli, P, Zheng, R.B, Haji-Ghassemi, O, Johal, A.R, Borisova, S, Lowary, T.L, Evans, S.V. | | Deposit date: | 2015-06-17 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | High Resolution Structures of the Human ABO(H) Blood Group Enzymes in Complex with Donor Analogs Reveal That the Enzymes Utilize Multiple Donor Conformations to Bind Substrates in a Stepwise Manner.
J.Biol.Chem., 290, 2015
|
|
4K3F
 
 | |
4K1R
 
 | |
2Y65
 
 | |
1X66
 
 | | Solution structure of the SAM_PNT-domain of the human friend LEUKEMIAINTEGRATION 1 transcription factor | | Descriptor: | Friend leukemia integration 1 transcription factor | | Authors: | Goroncy, A, Kigawa, T, Koshiba, S, Sato, M, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-17 | | Release date: | 2005-11-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SAM_PNT-domain of the human friend LEUKEMIAINTEGRATION 1 transcription factor
To be Published
|
|
1XYF
 
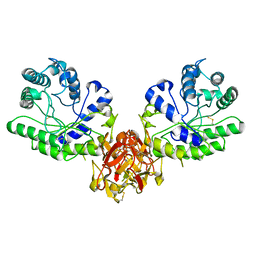 | | ENDO-1,4-BETA-XYLANASE FROM STREPTOMYCES OLIVACEOVIRIDIS | | Descriptor: | ENDO-1,4-BETA-XYLANASE | | Authors: | Fujimoto, Z, Mizuno, H, Kuno, A, Kusakabe, I. | | Deposit date: | 1999-05-11 | | Release date: | 2000-05-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Streptomyces olivaceoviridis E-86 beta-xylanase containing xylan-binding domain.
J.Mol.Biol., 300, 2000
|
|
4QOK
 
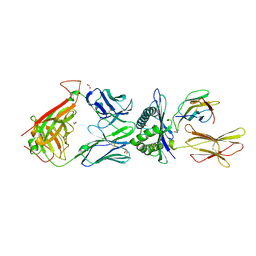 | | Structural basis for ineffective T-cell responses to MHC anchor residue improved heteroclitic peptides | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Rizkallah, P.J, Cole, D.K, Madura, F, Sewell, A.K. | | Deposit date: | 2014-06-20 | | Release date: | 2014-12-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for ineffective T-cell responses to MHC anchor residue-improved "heteroclitic" peptides.
Eur.J.Immunol., 45, 2015
|
|
2ZUV
 
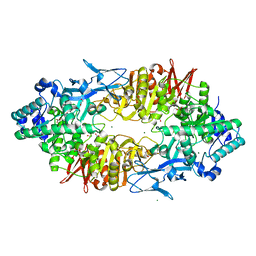 | | Crystal structure of Galacto-N-biose/Lacto-N-biose I phosphorylase in complex with GlcNAc, Ethylene glycol, and nitrate | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose, Lacto-N-biose phosphorylase, ... | | Authors: | Hidaka, M, Nishimoto, M, Kitaoka, M, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2008-10-28 | | Release date: | 2008-12-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structure of galacto-N-biose/lacto-N-biose I phosphorylase: A large deformation of a tim barrel scaffold
J.Biol.Chem., 284, 2009
|
|
5URN
 
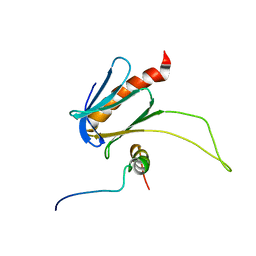 | | NMR structure of the complex between the PH domain of the Tfb1 subunit from TFIIH and the transactivation domain 1 of p65 | | Descriptor: | RNA polymerase II transcription factor B subunit 1, Transcription factor p65 | | Authors: | Lecoq, L, Omichinski, J.G, Raiola, L, Cyr, N, Chabot, P, Arseneault, G, Legault, P. | | Deposit date: | 2017-02-11 | | Release date: | 2017-03-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of interactions between transactivation domain 1 of the p65 subunit of NF-kappa B and transcription regulatory factors.
Nucleic Acids Res., 45, 2017
|
|
7K6O
 
 | | Crystal structure of PI3Kalpha inhibitor 10-5429 | | Descriptor: | (3S)-3-[4-(2-aminopyrimidin-5-yl)-2-(morpholin-4-yl)-5,6-dihydro-7H-pyrrolo[2,3-d]pyrimidin-7-yl]-N-methylpyrrolidine-1-sulfonamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Chen, P, Brooun, A, Deng, Y.L, Grodsky, N, Kaiser, S.E. | | Deposit date: | 2020-09-21 | | Release date: | 2021-01-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.738 Å) | | Cite: | Structure-Based Drug Design and Synthesis of PI3K alpha-Selective Inhibitor (PF-06843195).
J.Med.Chem., 64, 2021
|
|
2Y2K
 
 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) IN COMPLEX WITH AN ALKYL BORONATE (ZA5) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PENICILLIN-BINDING PROTEIN 1B, ... | | Authors: | Contreras-Martel, C, Amoroso, A, Woon, E, Zervosen, A, Inglis, S, Martins, A, Verlaine, O, Rydzik, A, Job, V, Luxen, A, Joris, B, Schofield, C, Dessen, A. | | Deposit date: | 2010-12-15 | | Release date: | 2011-08-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure-Guided Design of Cell Wall Biosynthesis Inhibitors that Overcome Beta-Lactam Resistance in Staphylococcus Aureus (Mrsa).
Acs Chem.Biol, 6, 2011
|
|
6JZP
 
 | |
3KU6
 
 | | Crystal structure of a H2N2 influenza virus hemagglutinin, 226L/228G | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xu, R, Wilson, I.A. | | Deposit date: | 2009-11-26 | | Release date: | 2010-01-19 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure, receptor binding, and antigenicity of influenza virus hemagglutinins from the 1957 H2N2 pandemic.
J.Virol., 84, 2010
|
|
1XOQ
 
 | | Catalytic Domain Of Human Phosphodiesterase 4D In Complex With Roflumilast | | Descriptor: | 1,2-ETHANEDIOL, 3-(CYCLOPROPYLMETHOXY)-N-(3,5-DICHLOROPYRIDIN-4-YL)-4-(DIFLUOROMETHOXY)BENZAMIDE, MAGNESIUM ION, ... | | Authors: | Card, G.L, England, B.P, Suzuki, Y, Fong, D, Powell, B, Lee, B, Luu, C, Tabrizizad, M, Gillette, S, Ibrahim, P.N, Artis, D.R, Bollag, G, Milburn, M.V, Kim, S.-H, Schlessinger, J, Zhang, K.Y.J. | | Deposit date: | 2004-10-06 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Basis for the Activity of Drugs that Inhibit Phosphodiesterases.
STRUCTURE, 12, 2004
|
|
4KIW
 
 | | Design and structural analysis of aromatic inhibitors of type II dehydroquinate dehydratase from Mycobacterium tuberculosis - compound 49e [5-[(3-nitrobenzyl)amino]benzene-1,3-dicarboxylic acid] | | Descriptor: | 3-dehydroquinate dehydratase, 5-[(3-nitrobenzyl)amino]benzene-1,3-dicarboxylic acid | | Authors: | Dias, M.V.B, Howard, N.G, Blundell, T.L, Abell, C. | | Deposit date: | 2013-05-02 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Design and Structural Analysis of Aromatic Inhibitors of Type II Dehydroquinase from Mycobacterium tuberculosis.
Chemmedchem, 10, 2015
|
|
6DEB
 
 | | Crystal Structure of Bifunctional Enzyme FolD-Methylenetetrahydrofolate Dehydrogenase/Cyclohydrolase in the Complex with Methotrexate from Campylobacter jejuni | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bifunctional protein FolD, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Maltseva, N, Grimshaw, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-05-11 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Bifunctional Enzyme FolD-Methylenetetrahydrofolate Dehydrogenase/Cyclohydrolase in the Complex with Methotrexate from Campylobacter jejuni
To Be Published
|
|
5XNC
 
 | | Crystal structure of the branched-chain polyamine synthase (BpsA) in complex with N4-aminopropylspermidine and 5-methylthioadenosine | | Descriptor: | 1,2-ETHANEDIOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mizohata, E, Tse, K.M, Fujita, J, Inoue, T. | | Deposit date: | 2017-05-22 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Active site geometry of a novel aminopropyltransferase for biosynthesis of hyperthermophile-specific branched-chain polyamine.
FEBS J., 284, 2017
|
|
4KPD
 
 | | Crystal Structure of Human Farnesyl Pyrophosphate Synthase (Y204F) Mutant Complexed with Mg, Risedronate and Isopentenyl Pyrophosphate | | Descriptor: | 1-HYDROXY-2-(3-PYRIDINYL)ETHYLIDENE BIS-PHOSPHONIC ACID, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Farnesyl pyrophosphate synthase, ... | | Authors: | Barnett, B.L, Tsoumpra, M.K, Muniz, J.R.C, Walter, R.L. | | Deposit date: | 2013-05-13 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structure of Human Farnesyl Pyrophosphate Synthase (Y204F) Mutant Complexed with Mg, Risedronate and Isopentenyl Pyrophosphate
To be Published
|
|
1ZAO
 
 | | Crystal Structure of A.fulgidus Rio2 Kinase Complexed With ATP and Manganese Ions | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Laronde-Leblanc, N, Guszczynski, T, Copeland, T, Wlodawer, A. | | Deposit date: | 2005-04-06 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Autophosphorylation of Archaeoglobus fulgidus Rio2 and crystal structures of its nucleotide-metal ion complexes.
Febs J., 272, 2005
|
|
3KQD
 
 | | Factor xa in complex with the inhibitor 1-(3-(5-oxo-4,5- dihydro-1h-1,2,4-triazol-3-yl)phenyl)-6-(2'-(pyrrolidin-1- ylmethyl)biphenyl-4-yl)-3-(trifluoromethyl)-5,6-dihydro- 1h-pyrazolo[3,4-c]pyridin-7(4h)-one | | Descriptor: | 1-(3-(5-OXO-4,5-DIHYDRO-1H-1,2,4-TRIAZOL-3-YL)PHENYL)-6-(2'-(PYRROLIDIN-1-YLMETHYL)BIPHENYL-4-YL)-3-(TRIFLUOROMETHYL)-5,6-DIHYDRO-1H-PYRAZOLO[3,4-C]PYRIDIN-7(4H)-ONE, SODIUM ION, factor Xa heavy chain, ... | | Authors: | Sheriff, S. | | Deposit date: | 2009-11-17 | | Release date: | 2010-02-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Phenyltriazolinones as potent factor Xa inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2Y5S
 
 | | Crystal structure of Burkholderia cenocepacia dihydropteroate synthase complexed with 7,8-dihydropteroate. | | Descriptor: | 1,2-ETHANEDIOL, 7,8-DIHYDROPTEROATE, CHLORIDE ION, ... | | Authors: | Morgan, R.E, Batot, G.O, Dement, J.M, Rao, V.A, Eadsforth, T.C, Hunter, W.N. | | Deposit date: | 2011-01-17 | | Release date: | 2011-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structures of Burkholderia Cenocepacia Dihydropteroate Synthase in the Apo-Form and Complexed with the Product 7,8-Dihydropteroate.
Bmc Struct.Biol., 11, 2011
|
|
3DMI
 
 | | Crystallization and Structural Analysis of Cytochrome c6 from the Diatom Phaeodactylum tricornutum at 1.5 A resolution | | Descriptor: | HEME C, MAGNESIUM ION, cytochrome c6 | | Authors: | Akazaki, H, Kawai, F, Hosokawa, M, Hama, T, Hirano, T, Lim, B.-K, Sakurai, N, Hakamata, W, Park, S.-Y, Nishio, T, Oku, T. | | Deposit date: | 2008-07-01 | | Release date: | 2009-03-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallization and structural analysis of cytochrome c(6) from the diatom Phaeodactylum tricornutum at 1.5 A resolution.
Biosci.Biotechnol.Biochem., 73, 2009
|
|
2Y4M
 
 | | MANNOSYLGLYCERATE SYNTHASE IN COMPLEX WITH GDP-Mannose | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GUANOSINE 5'-(TRIHYDROGEN DIPHOSPHATE), ... | | Authors: | Nielsen, M.M, Suits, M.D.L, Yang, M, Barry, C.S, Martinez-Fleites, C, Tailford, L.E, Flint, J.E, Davis, B.G, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2011-01-07 | | Release date: | 2011-02-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Substrate and Metal Ion Promiscuity in Mannosylglycerate Synthase.
J.Biol.Chem., 286, 2011
|
|
1SGC
 
 | |
1ONN
 
 | | IspC apo structure | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase | | Authors: | Steinbacher, S, Kaiser, J, Eisenreich, W, Huber, R, Bacher, A, Rohdich, F. | | Deposit date: | 2003-02-28 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of fosmidomycin action revealed by the complex with 2-C-methyl-D-erythritol
4-phosphate synthase (IspC). Implications for the catalytic mechanism and anti-malaria
drug development.
J.BIOL.CHEM., 278, 2003
|
|
