8P25
 
 | | Solution structure of a chimeric U2AF2 RRM2 / FUBP1 N-Box | | Descriptor: | Splicing factor U2AF 65 kDa subunit,Far upstream element-binding protein 1 | | Authors: | Hipp, C, Sattler, M. | | Deposit date: | 2023-05-14 | | Release date: | 2023-07-26 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | FUBP1 is a general splicing factor facilitating 3' splice site recognition and splicing of long introns.
Mol.Cell, 83, 2023
|
|
1U6F
 
 | | NMR solution structure of TcUBP1, a single RBD-unit from Trypanosoma cruzi | | Descriptor: | RNA-binding protein UBP1 | | Authors: | Volpon, L, D'orso, I, Frasch, A, Gehring, K. | | Deposit date: | 2004-07-29 | | Release date: | 2005-01-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structural Study of TcUBP1, a Single RRM Domain Protein from Trypanosoma cruzi: Contribution of a beta Hairpin to RNA Binding
Biochemistry, 44, 2005
|
|
6Y2D
 
 | | Crystal structure of the second KH domain of FUBP1 | | Descriptor: | Far upstream element-binding protein 1, GLYCEROL, SULFATE ION | | Authors: | Ni, X, Chaikuad, A, Joerger, A.C, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparative structural analyses and nucleotide-binding characterization of the four KH domains of FUBP1.
Sci Rep, 10, 2020
|
|
6Y2C
 
 | | Crystal structure of the third KH domain of FUBP1 | | Descriptor: | 1,2-ETHANEDIOL, Far upstream element-binding protein 1, ZINC ION | | Authors: | Ni, X, Joerger, A.C, Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Comparative structural analyses and nucleotide-binding characterization of the four KH domains of FUBP1.
Sci Rep, 10, 2020
|
|
6Y24
 
 | | Crystal structure of fourth KH domain of FUBP1 | | Descriptor: | 1,2-ETHANEDIOL, Far upstream element-binding protein 1 | | Authors: | Ni, X, Joerger, A.C, Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-14 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Comparative structural analyses and nucleotide-binding characterization of the four KH domains of FUBP1.
Sci Rep, 10, 2020
|
|
4LIJ
 
 | |
6RR0
 
 | | Crystal structure of the Sir4 H-BRCT domain in complex with Ubp10 pT123 peptide | | Descriptor: | CHLORIDE ION, Regulatory protein SIR4, Ubiquitin carboxyl-terminal hydrolase 10 | | Authors: | Deshpande, I, Keusch, J.J, Challa, K, Iesmantavicius, V, Gasser, S.M, Gut, H. | | Deposit date: | 2019-05-16 | | Release date: | 2019-09-18 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | The Sir4 H-BRCT domain interacts with phospho-proteins to sequester and repress yeast heterochromatin.
Embo J., 38, 2019
|
|
1J4W
 
 | |
1X4M
 
 | | Solution structure of KH domain in Far upstream element binding protein 1 | | Descriptor: | Far upstream element binding protein 1 | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of KH domain in Far upstream element binding protein 1
To be Published
|
|
1X4N
 
 | | Solution structure of KH domain in FUSE binding protein 1 | | Descriptor: | Far upstream element binding protein 1 | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of KH domain in FUSE binding protein 1
To be Published
|
|
2KXH
 
 | | Solution structure of the first two RRM domains of FIR in the complex with FBP Nbox peptide | | Descriptor: | Poly(U)-binding-splicing factor PUF60, peptide of Far upstream element-binding protein 1 | | Authors: | Cukier, C.D, Ramos, A, Hollingworth, D, Diaz-Moreno, I, Kelly, G. | | Deposit date: | 2010-05-05 | | Release date: | 2010-08-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of FIR-mediated c-myc transcriptional control.
Nat.Struct.Mol.Biol., 17, 2010
|
|
7AY1
 
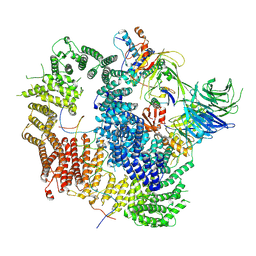 | | Cryo-EM structure of USP1-UAF1 bound to mono-ubiquitinated FANCD2, and FANCI | | Descriptor: | DNA (61-MER), Fanconi anemia group D2 protein, Fanconi anemia group I protein, ... | | Authors: | Rennie, M.L, Arkinson, C, Walden, H. | | Deposit date: | 2020-11-10 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of FANCD2 deubiquitination by USP1-UAF1.
Nat.Struct.Mol.Biol., 28, 2021
|
|
9FCI
 
 | | USP1 bound to KSQ-4279 and ubiquitin conjugated to FANCD2 (focused refinement) | | Descriptor: | 6-(4-cyclopropyl-6-methoxy-pyrimidin-5-yl)-1-[[4-[1-propan-2-yl-4-(trifluoromethyl)imidazol-2-yl]phenyl]methyl]pyrazolo[3,4-d]pyrimidine, Polyubiquitin-C, Ubiquitin carboxyl-terminal hydrolase 1, ... | | Authors: | Rennie, M.L, Gundogdu, M, Walden, H. | | Deposit date: | 2024-05-15 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and Biochemical Insights into the Mechanism of Action of the Clinical USP1 Inhibitor, KSQ-4279.
J.Med.Chem., 67, 2024
|
|
9FCJ
 
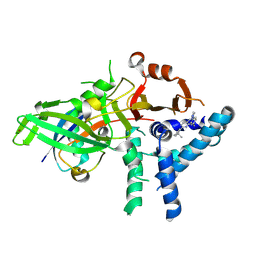 | | USP1 bound to ML323 and ubiquitin conjugated to FANCD2 (ordered subset, focused refinement) | | Descriptor: | 5-methyl-2-(2-propan-2-ylphenyl)-~{N}-[[4-(1,2,3-triazol-1-yl)phenyl]methyl]pyrimidin-4-amine, Polyubiquitin-C, Ubiquitin carboxyl-terminal hydrolase 1, ... | | Authors: | Rennie, M.L, Gundogdu, M, Walden, H. | | Deposit date: | 2024-05-15 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural and Biochemical Insights into the Mechanism of Action of the Clinical USP1 Inhibitor, KSQ-4279.
J.Med.Chem., 67, 2024
|
|
7ZH4
 
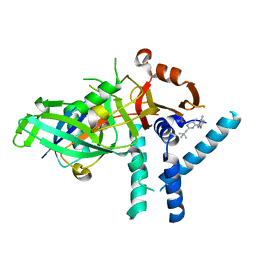 | | USP1 bound to ML323 and ubiquitin conjugated to FANCD2 (focused refinement) | | Descriptor: | 5-methyl-2-(2-propan-2-ylphenyl)-~{N}-[[4-(1,2,3-triazol-1-yl)phenyl]methyl]pyrimidin-4-amine, Ubiquitin carboxyl-terminal hydrolase 1, Ubiquitin-60S ribosomal protein L40, ... | | Authors: | Rennie, M.L, Walden, H. | | Deposit date: | 2022-04-05 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryo-EM reveals a mechanism of USP1 inhibition through a cryptic binding site.
Sci Adv, 8, 2022
|
|
7ZH3
 
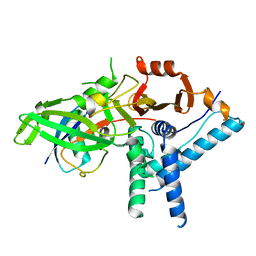 | |
8A9J
 
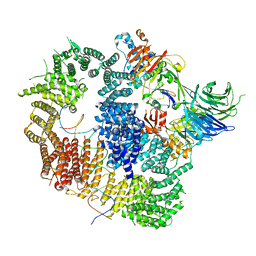 | |
8A9K
 
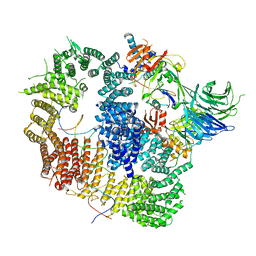 | | Cryo-EM structure of USP1-UAF1 bound to FANCI and mono-ubiquitinated FANCD2 with ML323 (consensus reconstruction) | | Descriptor: | 5-methyl-2-(2-propan-2-ylphenyl)-~{N}-[[4-(1,2,3-triazol-1-yl)phenyl]methyl]pyrimidin-4-amine, DNA (61-MER), Fanconi anemia group D2 protein, ... | | Authors: | Rennie, M.L, Walden, H. | | Deposit date: | 2022-06-28 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Cryo-EM reveals a mechanism of USP1 inhibition through a cryptic binding site.
Sci Adv, 8, 2022
|
|
3LMN
 
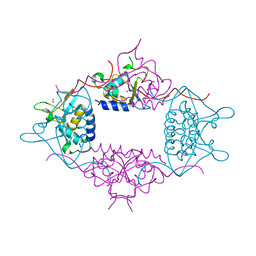 | | Oligomeric structure of the DUSP domain of human USP15 | | Descriptor: | ACETIC ACID, FORMIC ACID, Ubiquitin carboxyl-terminal hydrolase 15 | | Authors: | Walker, J.R, Asinas, A, Avvakumov, G.V, Alenkin, D, Weigelt, J, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Dhe-Paganon, S. | | Deposit date: | 2010-01-31 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of the Human Ubiquitin-Specific Protease 15 DUSP Domain
To be Published
|
|
3PPA
 
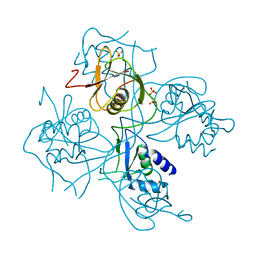 | | Structure of the Dusp-Ubl domains of Usp15 | | Descriptor: | CITRIC ACID, SULFATE ION, Ubiquitin carboxyl-terminal hydrolase 15 | | Authors: | Walker, J.R, Asinas, A.E, Dong, A, Weigelt, J, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-24 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of the Dusp-Ubl Domains of the Ubiquitin-Specific Protease 15
To be Published
|
|
4X3G
 
 | | Crystal structure of SIAH1 SINA domain in complex with a USP19 peptide | | Descriptor: | E3 ubiquitin-protein ligase SIAH1, Ubiquitin carboxyl-terminal hydrolase 19, ZINC ION | | Authors: | Walker, J.R, Dong, A, Zhang, Q, Huang, X, Li, Y, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-11-28 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal structure of SIAH1 SINA domain in complex with a USP19 peptide
To be published
|
|
5L8W
 
 | | Structure of USP12-UB-PRG/UAF1 | | Descriptor: | GLYCEROL, Polyubiquitin-B, Ubiquitin carboxyl-terminal hydrolase 12, ... | | Authors: | Dharadhar, S, Sixma, T. | | Deposit date: | 2016-06-08 | | Release date: | 2016-09-28 | | Last modified: | 2016-11-30 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | A conserved two-step binding for the UAF1 regulator to the USP12 deubiquitinating enzyme.
J.Struct.Biol., 196, 2016
|
|
6K7W
 
 | | Solution Structure of the CS1 Domain of USP19 | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 19 | | Authors: | Xue, W, Hu, H.Y. | | Deposit date: | 2019-06-10 | | Release date: | 2020-06-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Domain interactions reveal auto-inhibition of the deubiquitinating enzyme USP19 and its activation by HSP90 in the modulation of huntingtin aggregation.
Biochem.J., 477, 2020
|
|
8TH6
 
 | | Crystal Structure of the G3BP1 NTF2-like domain bound to USP10 peptide | | Descriptor: | 1,2-ETHANEDIOL, Ras GTPase-activating protein-binding protein 1, Ubiquitin carboxyl-terminal hydrolase 10 | | Authors: | Hughes, M.P, Taylor, J.P, Yang, Z. | | Deposit date: | 2023-07-14 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Interaction between host G3BP and viral nucleocapsid protein regulates SARS-CoV-2 replication and pathogenicity.
Cell Rep, 43, 2024
|
|
8WG5
 
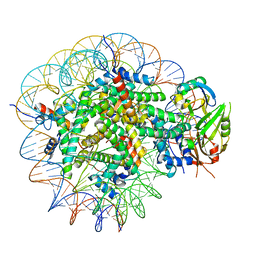 | | Cryo-EM structure of USP16 bound to H2AK119Ub nucleosome | | Descriptor: | DNA (147-MER), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | Authors: | Ai, H.S, He, Z.Z, Deng, Z.H, Liu, L. | | Deposit date: | 2023-09-20 | | Release date: | 2023-12-27 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural and mechanistic basis for nucleosomal H2AK119 deubiquitination by single-subunit deubiquitinase USP16.
Nat.Struct.Mol.Biol., 2024
|
|
