6PO0
 
 | |
3EWZ
 
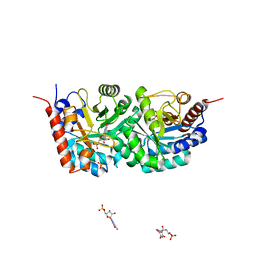 | |
1OKZ
 
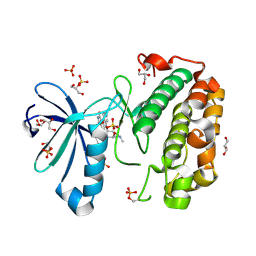 | | Structure of human PDK1 kinase domain in complex with UCN-01 | | Descriptor: | 3-PHOSPHOINOSITIDE DEPENDENT PROTEIN KINASE 1, 7-HYDROXYSTAUROSPORINE, GLYCEROL, ... | | Authors: | Komander, D, Kular, G.S, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2003-08-01 | | Release date: | 2004-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural Basis for Ucn-01 (7-Hydroxystaurosporine) Specificity and Pdk1 (3-Phosphoinositide-Dependent Protein Kinase-1) Inhibition
Biochem.J., 375, 2003
|
|
1AYN
 
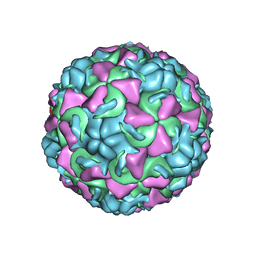 | | HUMAN RHINOVIRUS 16 COAT PROTEIN | | Descriptor: | HUMAN RHINOVIRUS 16 COAT PROTEIN, LAURIC ACID, MYRISTIC ACID, ... | | Authors: | Hadfield, A.T, Oliveira, M.A, Zhao, R, Rossmann, M.G. | | Deposit date: | 1997-11-06 | | Release date: | 1998-01-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of human rhinovirus 16.
Structure, 1, 1993
|
|
1AZB
 
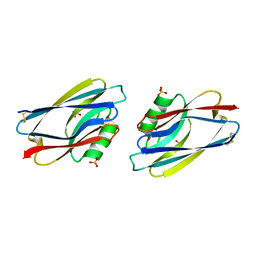 | |
1AZP
 
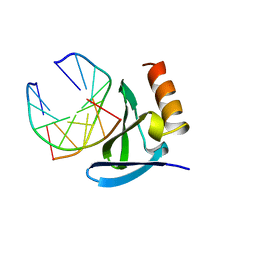 | | HYPERTHERMOPHILE CHROMOSOMAL PROTEIN SAC7D BOUND WITH KINKED DNA DUPLEX | | Descriptor: | DNA (5'-D(*GP*CP*GP*AP*TP*CP*GP*C)-3'), PROTEIN (HYPERTHERMOPHILE CHROMOSOMAL PROTEIN SAC7D) | | Authors: | Robinson, H, Gao, Y.-G, Mccrary, B.S, Edmondson, S.P, Shriver, J.W, Wang, A.H.-J. | | Deposit date: | 1997-11-19 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The hyperthermophile chromosomal protein Sac7d sharply kinks DNA.
Nature, 392, 1998
|
|
5GSY
 
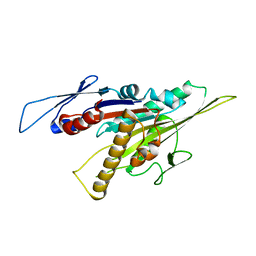 | | Kinesin-8 motor, KIF19A, in the nucleotide-free state complexed with GDP-taxol microtubule | | Descriptor: | Kinesin-like protein KIF19 | | Authors: | Morikawa, M, Nitta, R, Yajima, H, Shigematsu, H, Kikkawa, M, Hirokawa, N. | | Deposit date: | 2016-08-17 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Motility and microtubule depolymerization mechanisms of the Kinesin-8 motor, KIF19A
Elife, 5, 2016
|
|
1B3R
 
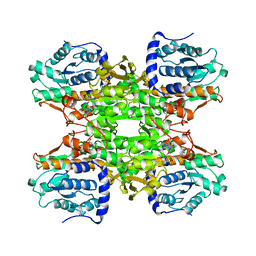 | | RAT LIVER S-ADENOSYLHOMOCYSTEIN HYDROLASE | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROTEIN (S-ADENOSYLHOMOCYSTEINE HYDROLASE) | | Authors: | Hu, Y, Komoto, J, Huang, Y, Takusagawa, F, Gomi, T, Ogawa, H, Takata, Y, Fujioka, M. | | Deposit date: | 1998-12-14 | | Release date: | 1998-12-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of S-adenosylhomocysteine hydrolase from rat liver.
Biochemistry, 38, 1999
|
|
1OKY
 
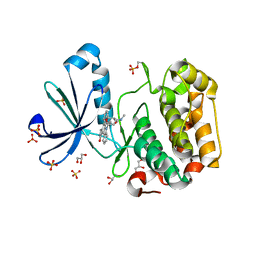 | | Structure of human PDK1 kinase domain in complex with staurosporine | | Descriptor: | 3-PHOSPHOINOSITIDE DEPENDENT PROTEIN KINASE 1, GLYCEROL, STAUROSPORINE, ... | | Authors: | Komander, D, Kular, G.S, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2003-08-01 | | Release date: | 2004-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Ucn-01 (7-Hydroxystaurosporine) Specificity and Pdk1 (3-Phosphoinositide-Dependent Protein Kinase-1) Inhibition
Biochem.J., 375, 2003
|
|
5GVQ
 
 | | Solution structure of the first RRM domain of human spliceosomal protein SF3b49 | | Descriptor: | Splicing factor 3B subunit 4 | | Authors: | Kuwasako, K, Nameki, N, Tsuda, K, Takahashi, M, Sato, A, Tochio, N, Inoue, M, Terada, T, Kigawa, T, Kobayashi, N, Shirouzu, M, Ito, T, Sakamoto, T, Wakamatsu, K, Guntert, P, Takahashi, S, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2016-09-06 | | Release date: | 2017-04-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the first RNA recognition motif domain of human spliceosomal protein SF3b49 and its mode of interaction with a SF3b145 fragment.
Protein Sci., 26, 2017
|
|
3EX6
 
 | | D312N mutant of human orotidyl-5'-monophosphate decarboxylase in complex with 6-azido-UMP, covalent adduct | | Descriptor: | CHLORIDE ION, GLYCEROL, Orotidine-5'-phosphate decarboxylase, ... | | Authors: | Heinrich, D, Diederichsen, U, Rudolph, M. | | Deposit date: | 2008-10-16 | | Release date: | 2009-04-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Lys314 is a nucleophile in non-classical reactions of orotidine-5'-monophosphate decarboxylase
Chemistry, 15, 2009
|
|
5JTN
 
 | | The structure of chaperone SecB in complex with unstructured proPhoA binding site c | | Descriptor: | Alkaline phosphatase, Protein-export protein SecB | | Authors: | Huang, C, Saio, T, Rossi, P, Kalodimos, C.G. | | Deposit date: | 2016-05-09 | | Release date: | 2016-08-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the antifolding activity of a molecular chaperone.
Nature, 537, 2016
|
|
6EWS
 
 | | Solution Structure of Rhabdopeptide NRPS Docking Domain Kj12A-NDD | | Descriptor: | NRPS Kj12A-NDD | | Authors: | Hacker, C, Cai, X, Kegler, C, Zhao, L, Weickhmann, A.K, Bode, H.B, Woehnert, J. | | Deposit date: | 2017-11-06 | | Release date: | 2018-10-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure-based redesign of docking domain interactions modulates the product spectrum of a rhabdopeptide-synthesizing NRPS.
Nat Commun, 9, 2018
|
|
1AW9
 
 | | STRUCTURE OF GLUTATHIONE S-TRANSFERASE III IN APO FORM | | Descriptor: | CADMIUM ION, GLUTATHIONE S-TRANSFERASE III | | Authors: | Neuefeind, T, Huber, R, Reinemer, P, Knaeblein, J. | | Deposit date: | 1997-10-13 | | Release date: | 1998-10-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cloning, sequencing, crystallization and X-ray structure of glutathione S-transferase-III from Zea mays var. mutin: a leading enzyme in detoxification of maize herbicides.
J.Mol.Biol., 274, 1997
|
|
1OL5
 
 | | Structure of Aurora-A 122-403, phosphorylated on Thr287, Thr288 and bound to TPX2 1-43 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RESTRICTED EXPRESSION PROLIFERATION ASSOCIATED PROTEIN 100, ... | | Authors: | Bayliss, R, Conti, E. | | Deposit date: | 2003-08-06 | | Release date: | 2003-10-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of Aurora-A Activation by Tpx2 at the Mitotic Spindle
Mol.Cell, 12, 2003
|
|
1OL7
 
 | |
1B7D
 
 | | NEUROTOXIN (TS1) FROM BRAZILIAN SCORPION TITYUS SERRULATUS | | Descriptor: | PHOSPHATE ION, PROTEIN (NEUROTOXIN TS1) | | Authors: | Polikarpov, I, Sanches Jr, M.S, Marangoni, S, Toyama, M.H, Teplyakov, A. | | Deposit date: | 1999-01-21 | | Release date: | 1999-07-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of neurotoxin Ts1 from Tityus serrulatus provides insights into the specificity and toxicity of scorpion toxins.
J.Mol.Biol., 290, 1999
|
|
1AP6
 
 | |
1WFC
 
 | | STRUCTURE OF APO, UNPHOSPHORYLATED, P38 MITOGEN ACTIVATED PROTEIN KINASE P38 (P38 MAP KINASE) THE MAMMALIAN HOMOLOGUE OF THE YEAST HOG1 PROTEIN | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE P38 | | Authors: | Wilson, K.P, Fitzgibbon, M.J, Caron, P.R, Griffith, J.P, Chen, W, Mccaffrey, P.G, Chambers, S.P, Su, M.S.-S. | | Deposit date: | 1996-09-13 | | Release date: | 1997-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of p38 mitogen-activated protein kinase.
J.Biol.Chem., 271, 1996
|
|
5GSE
 
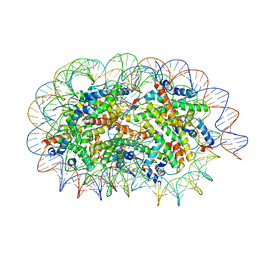 | | Crystal structure of unusual nucleosome | | Descriptor: | DNA (250-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Kato, D, Osakabe, A, Arimura, Y, Park, S.Y, Kurumizaka, H. | | Deposit date: | 2016-08-16 | | Release date: | 2017-05-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Crystal structure of the overlapping dinucleosome composed of hexasome and octasome
Science, 356, 2017
|
|
6SO5
 
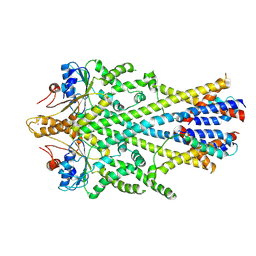 | | Homo sapiens WRB/CAML heterotetramer in complex with a TRC40 dimer | | Descriptor: | ATPase ASNA1, Calcium signal-modulating cyclophilin ligand, Tail-anchored protein insertion receptor WRB, ... | | Authors: | McDowell, M.A, Heimes, M, Wild, K, Flemming, D, Sinning, I. | | Deposit date: | 2019-08-29 | | Release date: | 2020-09-09 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural Basis of Tail-Anchored Membrane Protein Biogenesis by the GET Insertase Complex.
Mol.Cell, 80, 2020
|
|
1AXN
 
 | |
1PD8
 
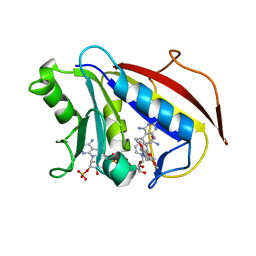 | | Analysis of Three Crystal Structure Determinations of a 5-Methyl-6-N-Methylanilino Pyridopyrimidine Antifolate Complex with Human Dihydrofolate Reductase | | Descriptor: | 2,4-DIAMINO-5-METHYL-6-[(3,4,5-TRIMETHOXY-N-METHYLANILINO)METHYL]PYRIDO[2,3-D]PYRIMIDINE, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Luft, J.R, Pangborn, W, Gangjee, A. | | Deposit date: | 2003-05-19 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Analysis of three crystal structure determinations of a 5-methyl-6-N-methylanilino pyridopyrimidine antifolate complex with human dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1PDB
 
 | | Analysis of Three Crystal Structure Determinations of a 5-Methyl-6-N-Methylanilino Pyridopyrimidine Antifolate Complex with Human Dihydrofolate Reductase | | Descriptor: | Dihydrofolate reductase | | Authors: | Cody, V, Luft, J.R, Pangborn, W, Gangjee, A. | | Deposit date: | 2003-05-19 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Analysis of three crystal structure determinations of a 5-methyl-6-N-methylanilino pyridopyrimidine antifolate complex with human dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1PE4
 
 | | SOLUTION STRUCTURE OF TOXIN CN12 FROM CENTRUROIDES NOXIUS ALFA SCORPION TOXIN ACTING ON SODIUM CHANNELS. NMR STRUCTURE | | Descriptor: | Neurotoxin Cn11 | | Authors: | Del Rio-Portilla, F, Hernandez-Marin, E, Pimienta, G, Coronas, F.V, Zamudio, F.Z, Rodrguez de la Vega, R.C, Wanke, E, Possani, L.D. | | Deposit date: | 2003-05-21 | | Release date: | 2004-05-25 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of Cn12, a novel peptide from the Mexican scorpion Centruroides noxius with a typical beta-toxin sequence but with alpha-like physiological activity.
Eur.J.Biochem., 271, 2004
|
|
