6YKS
 
 | | Neisseria gonorrhoeae Leucyl-tRNA Synthetase in Complex with Compound 11d | | Descriptor: | 1,2-ETHANEDIOL, Leucine--tRNA ligase, MAGNESIUM ION, ... | | Authors: | Pang, L, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2020-04-06 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Synthesis and structure-activity studies of novel anhydrohexitol-based Leucyl-tRNA synthetase inhibitors.
Eur.J.Med.Chem., 211, 2021
|
|
7BK4
 
 | | Crystal structure of RXRalpha ligand binding domain in complex with a fragment of the TIF2 coactivator | | Descriptor: | 6-[1-(3,5,5,8,8-PENTAMETHYL-5,6,7,8-TETRAHYDRONAPHTHALEN-2-YL)CYCLOPROPYL]PYRIDINE-3-CARBOXYLIC ACID, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | le Maire, A, Bourguet, W. | | Deposit date: | 2021-01-15 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights into the Interaction of the Intrinsically Disordered Co-activator TIF2 with Retinoic Acid Receptor Heterodimer (RXR/RAR).
J.Mol.Biol., 433, 2021
|
|
6YOW
 
 | |
8RUM
 
 | | Structure of Oceanobacillus iheyensis group II intron in the presence of Li+, Mg2+, and intronistat B | | Descriptor: | Domains 1-5, MAGNESIUM ION, ~{N}-(2-pyrrolidin-1-ylethyl)-2-[3,4,5-tris(oxidanyl)phenyl]carbonyl-1-benzofuran-5-carboxamide | | Authors: | Silvestri, I, Marcia, M. | | Deposit date: | 2024-01-31 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Targeting the conserved active site of splicing machines with specific and selective small molecule modulators.
Nat Commun, 15, 2024
|
|
7B91
 
 | |
5E9V
 
 | | Crystal structure of BRD9 bromodomain in complex with an indolizine ligand | | Descriptor: | 1,2-ETHANEDIOL, 1-[1-(imidazo[1,2-a]pyridin-5-yl)-7-(morpholin-4-yl)indolizin-3-yl]ethanone, Bromodomain-containing protein 9 | | Authors: | Tallant, C, Hay, D.A, Krojer, T, Nunez-Alonso, G, Picaud, S, Fedorov, O, Schofield, C.J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Knapp, S. | | Deposit date: | 2015-10-15 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of BRD9 bromodomain in complex with an indolizine ligand
To Be Published
|
|
6GT8
 
 | |
6DC3
 
 | | RSV prefusion F bound to RSD5 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab RSD5-Germline Heavy Chain, Fab RSD5-Germline Light Chain, ... | | Authors: | Battles, M.B, McLellan, J.S, Jones, H.J. | | Deposit date: | 2018-05-04 | | Release date: | 2019-07-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.501 Å) | | Cite: | Alternative conformations of a major antigenic site on RSV F.
Plos Pathog., 15, 2019
|
|
7BJ9
 
 | | Structure of Sfh-I with 2-Mercaptomethyl-thiazolidine L-anti-1a | | Descriptor: | (2~{S},4~{R})-2-ethoxycarbonyl-2-(sulfanylmethyl)-1,3-thiazolidine-4-carboxylic acid, Beta-lactamase, GLYCEROL, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2021-01-14 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.21000588 Å) | | Cite: | 2-Mercaptomethyl Thiazolidines (MMTZs) Inhibit All Metallo-beta-Lactamase Classes by Maintaining a Conserved Binding Mode.
Acs Infect Dis., 7, 2021
|
|
5EA1
 
 | | Crystal Structure of SMARCA4 bromodomain in complex with MPD | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Transcription activator BRG1 | | Authors: | Lolli, G, Caflisch, A. | | Deposit date: | 2015-10-15 | | Release date: | 2016-03-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-Throughput Fragment Docking into the BAZ2B Bromodomain: Efficient in Silico Screening for X-Ray Crystallography.
Acs Chem.Biol., 11, 2016
|
|
7BO6
 
 | | VDR complex with LCA derivative | | Descriptor: | (4R)-4-[(3R,5R,8R,9S,10S,13R,14S,17R)-10,13-dimethyl-3-(2-methyl-2-oxidanyl-propyl)-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl]pentanoic acid, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Rochel, N. | | Deposit date: | 2021-01-24 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Lithocholic acid-based design of noncalcemic vitamin D receptor agonists.
Bioorg.Chem., 111, 2021
|
|
5EA8
 
 | | Crystal Structure of Prefusion RSV F Glycoprotein Fusion Inhibitor Resistance Mutant D489Y | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, D(-)-TARTARIC ACID, Fusion glycoprotein F0, ... | | Authors: | Battles, M.B, McLellan, J.S, Arnoult, E, Roymans, D, Langedijk, J.P. | | Deposit date: | 2015-10-15 | | Release date: | 2015-12-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular mechanism of respiratory syncytial virus fusion inhibitors.
Nat.Chem.Biol., 12, 2016
|
|
6GFZ
 
 | | pVHL:EloB:EloC in complex with modified VH032 containing (3S,4S)-3-fluoro-4-hydroxyproline (ligand 14b) | | Descriptor: | (2~{R},3~{S},4~{S})-1-[(2~{S})-2-acetamido-3,3-dimethyl-butanoyl]-3-fluoranyl-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Gadd, M.S, Testa, A, Ciulli, A. | | Deposit date: | 2018-05-02 | | Release date: | 2018-07-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 3-Fluoro-4-hydroxyprolines: Synthesis, Conformational Analysis, and Stereoselective Recognition by the VHL E3 Ubiquitin Ligase for Targeted Protein Degradation.
J. Am. Chem. Soc., 140, 2018
|
|
7BFZ
 
 | | X-ray structure of human prostate-specific membrane antigen(PSMA) in complex with a inhibitor Glu-490 | | Descriptor: | (((S)-1-carboxy-5-((E)-2-cyano-3-(5-(1-(3-methoxy-3-oxopropyl)-1,2,3,4-tetrahydroquinolin-6-yl)thiophen-2-yl)acrylamido)pentyl)carbamoyl)-L-glutamic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rakhimbekova, A, Motlova, L, Barinka, C. | | Deposit date: | 2021-01-05 | | Release date: | 2021-08-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A prostate-specific membrane antigen activated molecular rotor for real-time fluorescence imaging.
Nat Commun, 12, 2021
|
|
6YQ0
 
 | | Promiscuous Reductase LugOII Catalyzes Keto-reduction at C1 during Lugdunomycin Biosynthesis | | Descriptor: | (3~{R})-8-methoxy-3-methyl-3-oxidanyl-2,4-dihydrobenzo[a]anthracene-1,7,12-trione, 1,2-ETHANEDIOL, Monooxygenase, ... | | Authors: | Xiao, X, Elsayed, S.S, Wu, C, van der Heul, H, Prota, A, Huang, J, Guo, R, Abrahams, J.P, van Wezel, G.P. | | Deposit date: | 2020-04-16 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Functional and Structural Insights into a Novel Promiscuous Ketoreductase of the Lugdunomycin Biosynthetic Pathway.
Acs Chem.Biol., 15, 2020
|
|
5E44
 
 | | Crystal structure of holo-FNR of A. fischeri | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FNR regulator, IRON/SULFUR CLUSTER | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2015-10-05 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The crystal structure of the global anaerobic transcriptional regulator FNR explains its extremely fine-tuned monomer-dimer equilibrium.
Sci Adv, 1, 2015
|
|
8SC7
 
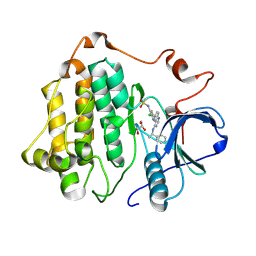 | |
6DD9
 
 | | Structure of mouse SYCP3, P1 form | | Descriptor: | Synaptonemal complex protein 3 | | Authors: | Rosenberg, S.C, Munoz, I.C, Uson, I, Corbett, K.D. | | Deposit date: | 2018-05-09 | | Release date: | 2018-08-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A conserved filamentous assembly underlies the structure of the meiotic chromosome axis.
Elife, 8, 2019
|
|
6YRB
 
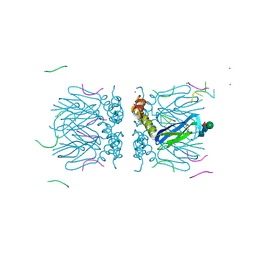 | | Crystal structure of the tetramerization domain of the glycoprotein Gn (Andes virus) at pH 7.5 | | Descriptor: | Envelope polyprotein, IODIDE ION, RNA (5'-D(*())-R(P*UP*UP*UP*())-3'), ... | | Authors: | Serris, A, Rey, F.A, Guardado-Calvo, P. | | Deposit date: | 2020-04-20 | | Release date: | 2020-10-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | The Hantavirus Surface Glycoprotein Lattice and Its Fusion Control Mechanism.
Cell, 183, 2020
|
|
6GP6
 
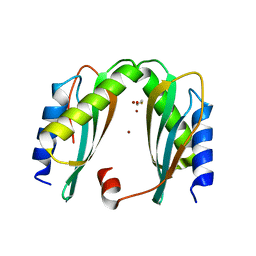 | | MamM CTD - Copper form | | Descriptor: | BETA-MERCAPTOETHANOL, COPPER (II) ION, Magnetosome protein MamM | | Authors: | Barber-Zucker, S, Zarivach, R. | | Deposit date: | 2018-06-05 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.146 Å) | | Cite: | MamM CTD - Copper form
To Be Published
|
|
7BIW
 
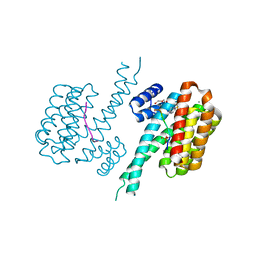 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-187 | | Descriptor: | 14-3-3 protein sigma, 4-(3,4-dihydro-2~{H}-quinoxalin-1-ylsulfonyl)benzaldehyde, CALCIUM ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-01-13 | | Release date: | 2021-09-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7BJB
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-044 | | Descriptor: | 14-3-3 protein sigma, 4-(4-methylphenyl)sulfonylmorpholine, CALCIUM ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-01-14 | | Release date: | 2021-09-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
6YLL
 
 | | Biochemical, Cellular and Structural Characterization of Novel ERK3 Inhibitors | | Descriptor: | Mitogen-activated protein kinase 6, ~{N}4-[3-(4-methoxyphenyl)-[1,2,3]triazolo[4,5-d]pyrimidin-5-yl]cyclohexane-1,4-diamine | | Authors: | Graedler, U. | | Deposit date: | 2020-04-07 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Biochemical, cellular and structural characterization of novel and selective ERK3 inhibitors.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6GPJ
 
 | | Crystal structure of human GDP-D-mannose 4,6-dehydratase in complex with GDP-4F-Man | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, GDP-mannose 4,6 dehydratase, ... | | Authors: | Pfeiffer, M, Krojer, T, Johansson, C, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Nidetzky, B, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-06 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A Parsimonious Mechanism of Sugar Dehydration by Human GDP-Mannose-4,6-dehydratase.
Acs Catalysis, 9, 2019
|
|
8S8A
 
 | | Human pyridoxal phosphatase in complex with 7,8-dihydroxyflavone without phosphate | | Descriptor: | 7,8-bis(oxidanyl)-2-phenyl-chromen-4-one, CHLORIDE ION, Chronophin, ... | | Authors: | Brenner, M, Gohla, A, Schindelin, H. | | Deposit date: | 2024-03-06 | | Release date: | 2024-06-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 7,8-Dihydroxyflavone is a direct inhibitor of human and murine pyridoxal phosphatase.
Elife, 13, 2024
|
|
