6VR8
 
 | | Structure of a pseudomurein peptide ligase type E from Methanothermus fervidus | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Mur ligase middle domain protein, ... | | Authors: | Carbone, V, Schofield, L.R, Sutherland-Smith, A.J, Ronimus, R.S, Subedi, B.P. | | Deposit date: | 2020-02-06 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterisation of methanogen pseudomurein cell wall peptide ligases homologous to bacterial MurE/F murein peptide ligases.
Microbiology (Reading, Engl.), 168, 2022
|
|
4X12
 
 | | JC Polyomavirus genotype 3 VP1 in complex with GD1b oligosaccharide | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Major capsid protein, ... | | Authors: | Stroh, L.J, Stehle, T. | | Deposit date: | 2014-11-24 | | Release date: | 2015-04-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Greater Affinity of JC Polyomavirus Capsid for alpha 2,6-Linked Lactoseries Tetrasaccharide c than for Other Sialylated Glycans Is a Major Determinant of Infectivity.
J.Virol., 89, 2015
|
|
6WL6
 
 | | Cocomplex structure of Deoxyhypusine synthase with inhibitor 6-[(2R)-1-AMINO-4-METHYLPENTAN-2-YL]-3-(PYRIDIN-3-YL)-4H,5H,6H,7H-THIENO[2,3-C]PYRIDIN-7-ONE | | Descriptor: | 6-[(2R)-1-amino-4-methylpentan-2-yl]-3-(pyridin-3-yl)-5,6-dihydrothieno[2,3-c]pyridin-7(4H)-one, Deoxyhypusine synthase | | Authors: | Klein, M.G, Ambrus-Aikelin, G. | | Deposit date: | 2020-04-18 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | New Series of Potent Allosteric Inhibitors of Deoxyhypusine Synthase.
Acs Med.Chem.Lett., 11, 2020
|
|
7MFC
 
 | |
8TRP
 
 | |
4WKF
 
 | | Crystal structure of human chitotriosidase-1 catalytic domain in complex with chitobiose (2.5mM) at 1.10 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitotriosidase-1 | | Authors: | Fadel, F, Zhao, Y, Cachau, R, Cousido-Siah, A, Ruiz, F.X, Harlos, K, Howard, E, Mitschler, A, Podjarny, A. | | Deposit date: | 2014-10-02 | | Release date: | 2015-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | New insights into the enzymatic mechanism of human chitotriosidase (CHIT1) catalytic domain by atomic resolution X-ray diffraction and hybrid QM/MM.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WTI
 
 | | CRYSTAL STRUCTURE OF HCV NS5B GENOTYPE 2A JFH-1 ISOLATE WITH S15G E86Q E87Q C223H V321I MUTATIONS IN COMPLEX WITH RNA TEMPLATE 5'-ACGG, RNA PRIMER 5'-PCC, MN2+, AND GDP | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Edwards, T.E, Appleby, T.C. | | Deposit date: | 2014-10-30 | | Release date: | 2015-02-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for RNA replication by the hepatitis C virus polymerase.
Science, 347, 2015
|
|
4TPY
 
 | | High throughput screening using acoustic droplet ejection to combine protein crystals and chemical libraries on crystallization plates at high density | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BENZAMIDINE, ... | | Authors: | Teplitsky, E, Joshi, K, Mullen, J.D, Soares, A.S. | | Deposit date: | 2014-06-10 | | Release date: | 2014-10-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High throughput screening using acoustic droplet ejection to combine protein crystals and chemical libraries on crystallization plates at high density.
J.Struct.Biol., 191, 2015
|
|
1E6V
 
 | | Methyl-coenzyme M reductase from Methanopyrus kandleri | | Descriptor: | 1-THIOETHANESULFONIC ACID, Coenzyme B, FACTOR 430, ... | | Authors: | Grabarse, W, Ermler, U. | | Deposit date: | 2000-08-23 | | Release date: | 2000-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Comparison of Three Methyl-Coenzyme M Reductases from Phylogenetically Distant Organisms: Unusual Amino Acid Modification, Conservation and Adaptation
J.Mol.Biol., 303, 2000
|
|
4WYD
 
 | | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase from Mycobacterium tuberculosis complexed with a fragment from DSF screening | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, ... | | Authors: | Dai, R, Finzel, B.C, Geders, T.W. | | Deposit date: | 2014-11-17 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Fragment-Based Exploration of Binding Site Flexibility in Mycobacterium tuberculosis BioA.
J.Med.Chem., 58, 2015
|
|
6Y87
 
 | |
4S29
 
 | | Crystal structure of Arabidopsis thaliana ThiC with bound imidazole ribonucleotide and Fe | | Descriptor: | 1,4-BUTANEDIOL, 1-(5-O-phosphono-beta-D-ribofuranosyl)-1H-imidazole, FE (II) ION, ... | | Authors: | Fenwick, M.K, Mehta, A.P, Zhang, Y, Abdelwahed, S, Begley, T.P, Ealick, S.E. | | Deposit date: | 2015-01-19 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.382 Å) | | Cite: | Non-canonical active site architecture of the radical SAM thiamin pyrimidine synthase.
Nat Commun, 6
|
|
4S3G
 
 | | Structure of the F249X mutant of Phosphatidylinositol-specific phospholipase C from Staphylococcus aureus | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1-phosphatidylinositol phosphodiesterase, ACETATE ION | | Authors: | He, T, Gershenson, A, Eyles, S.J, Gao, J, Roberts, M.F. | | Deposit date: | 2015-01-26 | | Release date: | 2015-07-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fluorinated Aromatic Amino Acids Distinguish Cation-pi Interactions from Membrane Insertion.
J.Biol.Chem., 290, 2015
|
|
6YH7
 
 | | Crystal structure of chimeric carbonic anhydrase XII with 3-[(1S)-2,3-Dihydro-1H-inden-1-ylamino]-2,5,6-trifluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide | | Descriptor: | 3-[(1S)-2,3-dihydro-1H-inden-1-ylamino]-2,5,6-trifluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2020-03-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Switching the Inhibitor-Enzyme Recognition Profile via Chimeric Carbonic Anhydrase XII.
Chemistryopen, 10, 2021
|
|
7MCJ
 
 | |
6YH9
 
 | | Crystal structure of chimeric carbonic anhydrase XII with 2,3,6-trifluoro-5-{[(1R,2S)-2-hydroxy-1,2-diphenylethyl]amino}-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide | | Descriptor: | 2,3,6-trifluoro-5-{[(1R,2S)-2-hydroxy-1,2-diphenylethyl]amino}-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2020-03-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Switching the Inhibitor-Enzyme Recognition Profile via Chimeric Carbonic Anhydrase XII.
Chemistryopen, 10, 2021
|
|
8FXJ
 
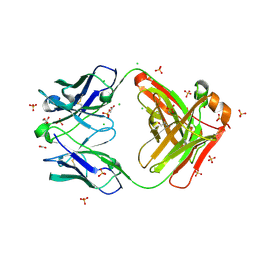 | | Crystal structure of Fab460 | | Descriptor: | ACETATE ION, CHLORIDE ION, Fab460, ... | | Authors: | Tan, K, Kim, M, Reinherz, E.L. | | Deposit date: | 2023-01-24 | | Release date: | 2023-10-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inadequate structural constraint on Fab approach rather than paratope elicitation limits HIV-1 MPER vaccine utility.
Nat Commun, 14, 2023
|
|
4WMW
 
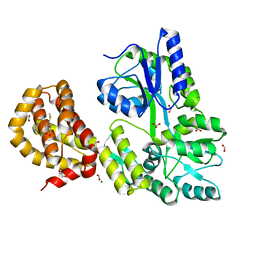 | | The structure of MBP-MCL1 bound to ligand 5 at 1.9A | | Descriptor: | 1,2-ETHANEDIOL, 2-hydroxy-5-(methylsulfanyl)benzoic acid, FORMIC ACID, ... | | Authors: | Clifton, M.C, Dranow, D.M. | | Deposit date: | 2014-10-09 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Maltose-Binding Protein Fusion Construct Yields a Robust Crystallography Platform for MCL1.
Plos One, 10, 2015
|
|
6OTN
 
 | | Crystal Structure of an N-terminal Fragment of Cancer Associated Tropomyosin 3.1 (Tpm3.1) | | Descriptor: | SULFATE ION, Tropomyosin alpha-3 chain | | Authors: | Rynkiewicz, M.J, Ghosh, A, Lehman, W.J, Janco, M, Gunning, P.W. | | Deposit date: | 2019-05-03 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular integration of the anti-tropomyosin compound ATM-3507 into the coiled coil overlap region of the cancer-associated Tpm3.1.
Sci Rep, 9, 2019
|
|
6WNP
 
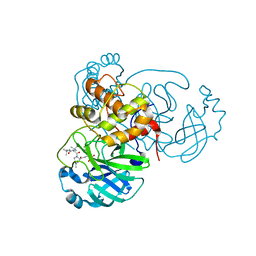 | |
4TT7
 
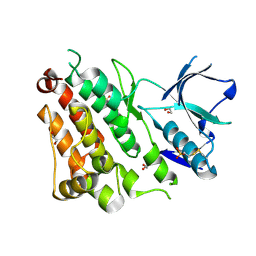 | | Crystal structure of human ALK with a covalent modification | | Descriptor: | 1,2-ETHANEDIOL, 1-THIOETHANESULFONIC ACID, ALK tyrosine kinase receptor | | Authors: | Badger, J, Sridhar, V, Chie-Leon, B, Nienaber, V.L, Hausheer, F.H. | | Deposit date: | 2014-06-19 | | Release date: | 2015-02-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel covalent modification of human anaplastic lymphoma kinase (ALK) and potentiation of crizotinib-mediated inhibition of ALK activity by BNP7787.
Onco Targets Ther, 8, 2015
|
|
6IJE
 
 | | Crystal structure of the type VI amidase immunity (Tai4) from Agrobacterium tumefaciens | | Descriptor: | 1,2-ETHANEDIOL, Tai4 | | Authors: | Fukuhara, S, Nakane, T, Yamashita, K, Ishii, R, Ishitani, R, Nureki, O. | | Deposit date: | 2018-10-09 | | Release date: | 2018-12-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of the Agrobacterium tumefaciens type VI effector-immunity complex.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
4S25
 
 | | Crystal structure of Arabidopsis thaliana ThiC with bound imidazole ribonucleotide, S-adenosylhomocysteine, Fe4S4 cluster and Zn (trigonal crystal form) | | Descriptor: | 1,4-BUTANEDIOL, 1-(5-O-phosphono-beta-D-ribofuranosyl)-1H-imidazole, CHLORIDE ION, ... | | Authors: | Fenwick, M.K, Mehta, A.P, Zhang, Y, Abdelwahed, S, Begley, T.P, Ealick, S.E. | | Deposit date: | 2015-01-19 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Non-canonical active site architecture of the radical SAM thiamin pyrimidine synthase.
Nat Commun, 6
|
|
4WMS
 
 | | STRUCTURE OF APO MBP-MCL1 AT 1.9A | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Clifton, M.C, Dranow, D.M. | | Deposit date: | 2014-10-09 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Maltose-Binding Protein Fusion Construct Yields a Robust Crystallography Platform for MCL1.
Plos One, 10, 2015
|
|
4TKX
 
 | | Structure of Protease | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, LEAD (II) ION, ... | | Authors: | Gorman, M.A, Parker, M.W. | | Deposit date: | 2014-05-28 | | Release date: | 2014-12-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the lysine specific protease Kgp from Porphyromonas gingivalis, a target for improved oral health.
Protein Sci., 24, 2015
|
|
