6Z2K
 
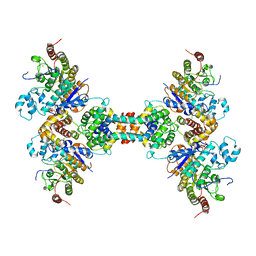 | | The structure of the tetrameric HDAC1/MIDEAS/DNTTIP1 MiDAC deacetylase complex | | Descriptor: | Deoxynucleotidyltransferase terminal-interacting protein 1, Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Fairall, L, Saleh, A, Ragan, T.J, Millard, C.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-05-16 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | The MiDAC histone deacetylase complex is essential for embryonic development and has a unique multivalent structure.
Nat Commun, 11, 2020
|
|
4OME
 
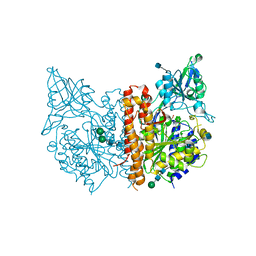 | | X-ray structure of human glutamate carboxypeptidase II in complex with DCCBL, a urea based inhibitor with distal carborane moiety | | Descriptor: | (S)-2-(3-((S)-1-carboxy-5-(1,2-dicarba-closo-dodecarboranylamido) pentyl)ureido)pentanedioic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Barinka, C, Ptacek, J, Novakova, Z, Byun, Y. | | Deposit date: | 2014-01-27 | | Release date: | 2015-02-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | X-ray structure of human glutamate carboxypeptidase II in complex with DCCBL, a urea based inhibitor with distal carborane moiety
To be Published
|
|
6WC5
 
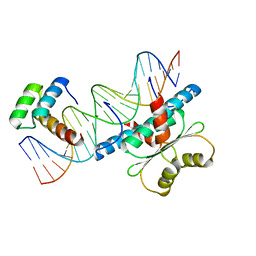 | | Crystal Structure of a Ternary MEF2B/NKX2-5/myocardin enhancer DNA Complex | | Descriptor: | Homeobox protein Nkx-2.5, Myocardin enhancer DNA, Myocyte-specific enhancer factor 2B | | Authors: | Chen, L, Lei, X. | | Deposit date: | 2020-03-29 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structures of Ternary Complexes of MEF2 and NKX2-5 Bound to DNA Reveal a Disease Related Protein-Protein Interaction Interface.
J.Mol.Biol., 432, 2020
|
|
4OC3
 
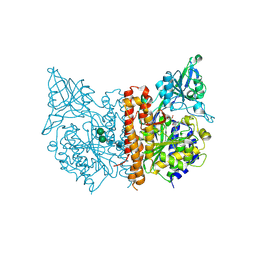 | | X-ray structure of of human glutamate carboxypeptidase II (GCPII) in a complex with CFIBzL, a urea-based inhibitor N~2~-{[(1S)-1-carboxy-2-(furan-2-yl)ethyl]carbamoyl}-N~6~-(4-iodobenzoyl)-L-lysine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pavlicek, J, Ptacek, J, Cerny, J, Byun, Y, Skultetyova, L, Pomper, M, Lubkowski, J, Barinka, C. | | Deposit date: | 2014-01-08 | | Release date: | 2014-05-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural characterization of P1'-diversified urea-based inhibitors of glutamate carboxypeptidase II.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
6WG3
 
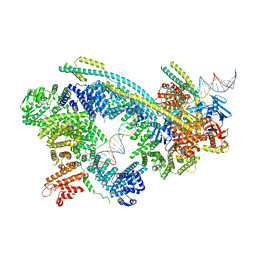 | | Cryo-EM structure of human Cohesin-NIPBL-DNA complex | | Descriptor: | Cohesin subunit SA-1, DNA (51-MER), Double-strand-break repair protein rad21 homolog, ... | | Authors: | Shi, Z.B, Gao, H, Bai, X.C, Yu, H. | | Deposit date: | 2020-04-04 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Cryo-EM structure of the human cohesin-NIPBL-DNA complex.
Science, 368, 2020
|
|
4ONG
 
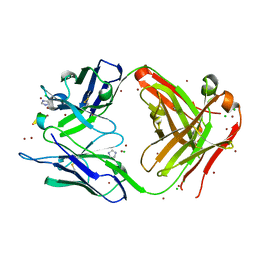 | | Fab fragment of 3D6 in complex with amyloid beta 1-40 | | Descriptor: | 3D6 FAB ANTIBODY HEAVY CHAIN, 3D6 FAB ANTIBODY LIGHT CHAIN, Amyloid beta A4 protein, ... | | Authors: | Feinberg, H, Saldanha, J.W, Diep, L, Goel, A, Widom, A, Veldman, G.M, Weis, W.I, Schenk, D, Basi, G.S. | | Deposit date: | 2014-01-28 | | Release date: | 2014-06-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure reveals conservation of amyloid-beta conformation recognized by 3D6 following humanization to bapineuzumab.
Alzheimers Res Ther, 6, 2014
|
|
6WCQ
 
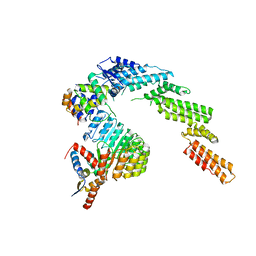 | | Structure of a substrate-bound DQC ubiquitin ligase | | Descriptor: | Cullin-1, F-box/LRR-repeat protein 17, Kelch-like ECH-associated protein 1, ... | | Authors: | Mena, E.L, Jevtic, P, Greber, B.J, Gee, C.L, Lew, B.G, Akopian, D, Nogales, E, Kuriyan, J, Rape, M. | | Deposit date: | 2020-03-31 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Structural basis for dimerization quality control.
Nature, 586, 2020
|
|
6WD7
 
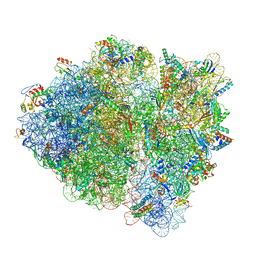 | | Cryo-EM of elongating ribosome with EF-Tu*GTP elucidates tRNA proofreading (Cognate Structure II-D) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Korostelev, A.A. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM of elongating ribosome with EF-Tu•GTP elucidates tRNA proofreading.
Nature, 584, 2020
|
|
6WCB
 
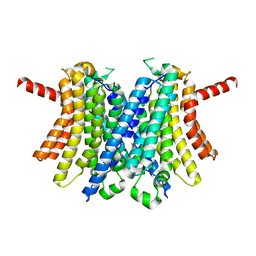 | | Human open state TMEM175 in CsCl | | Descriptor: | CESIUM ION, Endosomal/lysosomal potassium channel TMEM175 | | Authors: | Oh, S, Paknejad, N, Hite, R.K. | | Deposit date: | 2020-03-30 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Gating and selectivity mechanisms for the lysosomal K + channel TMEM175.
Elife, 9, 2020
|
|
6WDG
 
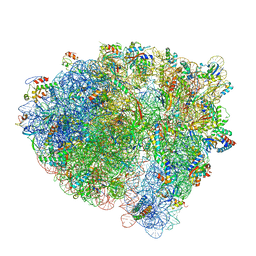 | | Cryo-EM of elongating ribosome with EF-Tu*GTP elucidates tRNA proofreading (Cognate Structure VI-B) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Korostelev, A.A. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM of elongating ribosome with EF-Tu•GTP elucidates tRNA proofreading.
Nature, 584, 2020
|
|
6WD1
 
 | | Cryo-EM of elongating ribosome with EF-Tu*GTP elucidates tRNA proofreading (Cognate Structure I-B) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Korostelev, A.A. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM of elongating ribosome with EF-Tu•GTP elucidates tRNA proofreading.
Nature, 584, 2020
|
|
6WHA
 
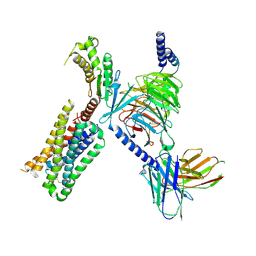 | | HTR2A bound to 25-CN-NBOH in complex with a mini-Galpha-q protein, beta/gamma subunits and an active-state stabilizing single-chain variable fragment (scFv16) obtained by cryo-electron microscopy (cryoEM) | | Descriptor: | 4-(2-{[(2-hydroxyphenyl)methyl]amino}ethyl)-2,5-dimethoxybenzonitrile, G subunit q (Gi2-mini-Gq chimeric), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Skiniotis, G, Roth, B, Kim, K, Panova, O. | | Deposit date: | 2020-04-07 | | Release date: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structure of a Hallucinogen-Activated Gq-Coupled 5-HT2A Serotonin Receptor
Cell(Cambridge,Mass.), 182, 2020
|
|
5VTG
 
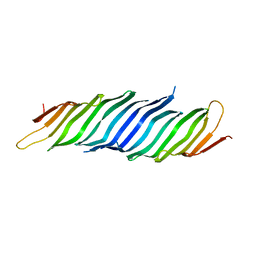 | |
6WHR
 
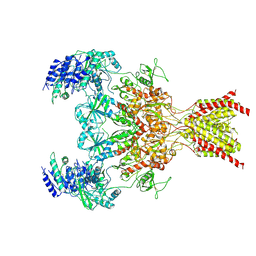 | | GluN1b-GluN2B NMDA receptor in non-active 2 conformation at 4 angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Chou, T, Tajima, N, Furukawa, H. | | Deposit date: | 2020-04-08 | | Release date: | 2020-08-05 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
4OF9
 
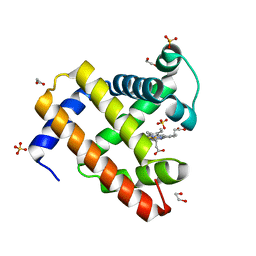 | | Structure of K42N variant of sperm whale myoglobin | | Descriptor: | 1,2-ETHANEDIOL, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lebioda, L, Wang, C, Lovelace, L.L. | | Deposit date: | 2014-01-14 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.241 Å) | | Cite: | Structures of K42N and K42Y sperm whale myoglobins point to an inhibitory role of distal water in peroxidase activity.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6WI9
 
 | | Human secretin receptor Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Belousoff, M.J, Khoshouei, M. | | Deposit date: | 2020-04-09 | | Release date: | 2020-08-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure and dynamics of the active Gs-coupled human secretin receptor.
Nat Commun, 11, 2020
|
|
6WJ3
 
 | |
6WDH
 
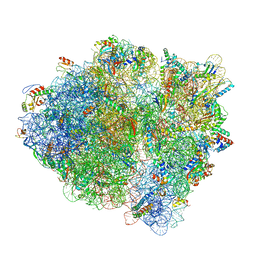 | | Cryo-EM of elongating ribosome with EF-Tu*GTP elucidates tRNA proofreading (Non-cognate Structure IV-B1) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Demo, G, Korostelev, A.A. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM of elongating ribosome with EF-Tu•GTP elucidates tRNA proofreading.
Nature, 584, 2020
|
|
5VLH
 
 | | Short PCSK9 delta-P' complex with peptide Pep1 | | Descriptor: | ACE-THR-VAL-PHE-THR-SER-TRP-GLU-GLU-TYR-LEU-ASP-TRP-VAL-NH2, CALCIUM ION, CYS-ARG-LEU-PRO-TRP-ASN-LEU-GLN-ARG-ILE-GLY-LEU-PRO-CYS, ... | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2017-04-25 | | Release date: | 2017-08-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Discovery of a cryptic peptide-binding site on PCSK9 and design of antagonists.
Nat. Struct. Mol. Biol., 24, 2017
|
|
6WKF
 
 | | Crystal structure of pentalenene synthase mutant F76Y complexed with 12,13-difluorofarnesyl diphosphate | | Descriptor: | (2E,6E)-12-fluoro-11-(fluoromethyl)-3,7-dimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, Pentalenene synthase | | Authors: | Prem Kumar, R, Matos, J.O, Oprian, D.D. | | Deposit date: | 2020-04-16 | | Release date: | 2020-08-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism Underlying Anti-Markovnikov Addition in the Reaction of Pentalenene Synthase.
Biochemistry, 59, 2020
|
|
5VLL
 
 | | Short PCSK9 delta-P' complex with peptide Pep3 | | Descriptor: | ACE-THR-VAL-PHE-THR-SER-TRP-GLU-GLU-TYR-LEU-ASP-TRP-VAL-NH2, CALCIUM ION, CYS-PHE-ILE-PRO-TRP-ASN-LEU-GLN-ARG-ILE-GLY-LEU-LEU-CYS, ... | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2017-04-25 | | Release date: | 2017-08-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Discovery of a cryptic peptide-binding site on PCSK9 and design of antagonists.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5VLQ
 
 | | Structure of the TTLL3 Glycylase | | Descriptor: | LOC100158544 protein, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SULFATE ION | | Authors: | Garnham, C.P, Yu, I, Li, Y, Roll-Mecak, A. | | Deposit date: | 2017-04-25 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.285 Å) | | Cite: | Crystal structure of tubulin tyrosine ligase-like 3 reveals essential architectural elements unique to tubulin monoglycylases.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4OR9
 
 | | Crystal structure of human calcineurin | | Descriptor: | CALCIUM ION, Calcineurin subunit B type 1, FE (III) ION, ... | | Authors: | Li, S.J, Wang, J, Wu, J.W, Wang, Z.X. | | Deposit date: | 2014-02-11 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Cooperative autoinhibition and multi-level activation mechanisms of calcineurin
To be Published
|
|
6WW5
 
 | | Structure of VcINDY-Na-Fab84 in nanodisc | | Descriptor: | 1,2-DIHEXANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, DASS family sodium-coupled anion symporter, Fab84 Heavy Chain, ... | | Authors: | Sauer, D.B, Marden, J, Song, J.M, Koide, A, Koide, S, Wang, D.N. | | Deposit date: | 2020-05-07 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural basis for the reaction cycle of DASS dicarboxylate transporters.
Elife, 9, 2020
|
|
5VNK
 
 | |
