7DMU
 
 | |
7DS7
 
 | | The Crystal Structure of Leaf-branch compost cutinase from Biortus. | | Descriptor: | CITRIC ACID, GLYCEROL, IMIDAZOLE, ... | | Authors: | Wang, F, Lv, Z, Cheng, W, Lin, D, Chu, F, Xu, X, Tan, J. | | Deposit date: | 2020-12-30 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Crystal Structure of Leaf-branch compost cutinase from Biortus.
To Be Published
|
|
7D92
 
 | | Crystal Structure of the Na+,K+-ATPase in the E2P state with bound Mg2+ and anthroylouabain (P4(3)2(1)2 symmetry) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kanai, R, Cornelius, F, Ogawa, H, Motoyama, K, Vilsen, B, Toyoshima, C. | | Deposit date: | 2020-10-12 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Binding of cardiotonic steroids to Na + ,K + -ATPase in the E2P state.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8UZ6
 
 | |
8V0K
 
 | |
2HZP
 
 | | Crystal Structure of Homo Sapiens Kynureninase | | Descriptor: | Kynureninase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Lima, S, Khristoforov, R, Momany, C, Phillips, R.S. | | Deposit date: | 2006-08-09 | | Release date: | 2006-11-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Homo sapiens Kynureninase.
Biochemistry, 46, 2007
|
|
2I1K
 
 | |
2IA8
 
 | | Kinetic and Crystallographic Studies of a Redesigned Manganese-Binding Site in Cytochrome c Peroxidase | | Descriptor: | Cytochrome c peroxidase, mitochondrial, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Pfister, T, Mirarefi, A.Y, Gengenbach, A.J, Zhao, X, Conaster, C.D.N, Gao, Y.G, Robinson, H, Zukoski, C.F, Wang, A.H.J, Lu, Y. | | Deposit date: | 2006-09-07 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Kinetic and crystallographic studies of a redesigned manganese-binding site in cytochrome c peroxidase
J.Biol.Inorg.Chem., 12, 2007
|
|
7D7S
 
 | |
7DA5
 
 | | Cryo-EM structure of the human MCT1 D309N mutant in complex with Basigin-2 in the inward-open conformation. | | Descriptor: | Basigin, Monocarboxylate transporter 1 | | Authors: | Wang, N, Jiang, X, Zhang, S, Zhu, A, Yuan, Y, Lei, J, Yan, C. | | Deposit date: | 2020-10-14 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of human monocarboxylate transporter 1 inhibition by anti-cancer drug candidates.
Cell, 184, 2021
|
|
8V0I
 
 | |
8SW0
 
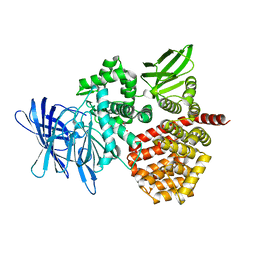 | | Puromycin sensitive aminopeptidase | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Puromycin-sensitive aminopeptidase, ... | | Authors: | Rodgers, D.W, Sampath, S. | | Deposit date: | 2023-05-17 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structure of puromycin-sensitive aminopeptidase and polyglutamine binding.
Plos One, 18, 2023
|
|
2I7B
 
 | |
8T2V
 
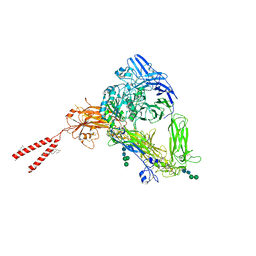 | | Cryo-EM Structures of Full-length Integrin alphaIIbbeta3 in Native Lipids | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Adair, B, Xiong, J.P, Yeager, M, Arnaout, M.A. | | Deposit date: | 2023-06-06 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of full-length integrin alpha IIb beta 3 in native lipids.
Nat Commun, 14, 2023
|
|
8SPH
 
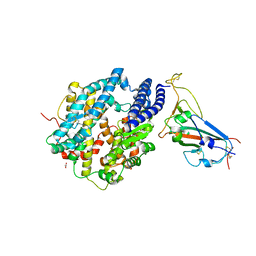 | | Crystal structure of chimeric omicron RBD (strain XBB.1) complexed with human ACE2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, W, Shi, K, Aihara, H, Li, F. | | Deposit date: | 2023-05-03 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural evolution of SARS-CoV-2 omicron in human receptor recognition.
J.Virol., 97, 2023
|
|
7D93
 
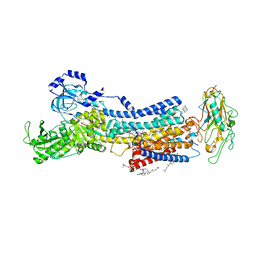 | | Crystal Structure of the Na+,K+-ATPase in the E2P state with bound Mg2+ and anthroylouabain (P2(1)2(1)2(1) symmetry) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kanai, R, Cornelius, F, Ogawa, H, Motoyama, K, Vilsen, B, Toyoshima, C. | | Deposit date: | 2020-10-12 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Binding of cardiotonic steroids to Na + ,K + -ATPase in the E2P state.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7DDI
 
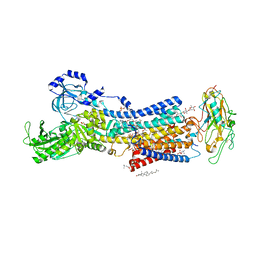 | | Crystal structures of Na+,K+-ATPase in complex with digitoxin | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ogawa, H, Cornelius, F, Kanai, R, Motoyama, K, Vilsen, B, Toyoshima, C. | | Deposit date: | 2020-10-29 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.72 Å) | | Cite: | Binding of cardiotonic steroids to Na + ,K + -ATPase in the E2P state.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7DI8
 
 | |
8SPI
 
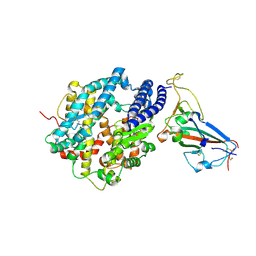 | | Crystal structure of chimeric omicron RBD (strain XBB.1.5) complexed with human ACE2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, W, Shi, K, Aihara, H, Li, F. | | Deposit date: | 2023-05-03 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structural evolution of SARS-CoV-2 omicron in human receptor recognition.
J.Virol., 97, 2023
|
|
7D8Z
 
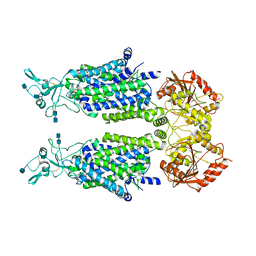 | | human potassium-chloride co-transporter KCC2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, potassium-chloride cotransporter 2 | | Authors: | Xie, Y, Chang, S, Zhao, C, Ye, S, Guo, J. | | Deposit date: | 2020-10-11 | | Release date: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures and an activation mechanism of human potassium-chloride cotransporters.
Sci Adv, 6, 2020
|
|
2IC0
 
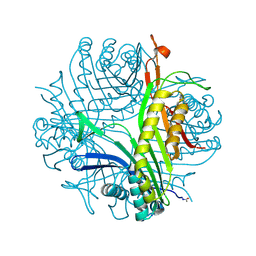 | | Urate oxidase under 2.0 MPa pressure of xenon | | Descriptor: | 8-AZAXANTHINE, SODIUM ION, Uricase, ... | | Authors: | Colloc'h, N, Retailleau, P, Sopkova-de Oliveira Santos, J, Abraini, J.H, Prange, T. | | Deposit date: | 2006-09-12 | | Release date: | 2007-02-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Protein Crystallography under Xenon and Nitrous Oxide Pressure: Comparison with In Vivo Pharmacology Studies and Implications for the Mechanism of Inhaled Anesthetic Action
Biophys.J., 92, 2007
|
|
7D90
 
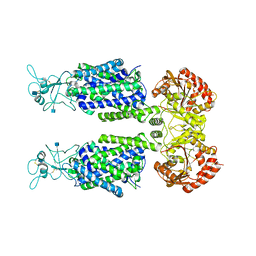 | | human potassium-chloride co-transporter KCC3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, potassium-chloride cotransporter 3 | | Authors: | Xie, Y, Chang, S, Zhao, C, Ye, S, Guo, J. | | Deposit date: | 2020-10-12 | | Release date: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures and an activation mechanism of human potassium-chloride cotransporters.
Sci Adv, 6, 2020
|
|
7DDK
 
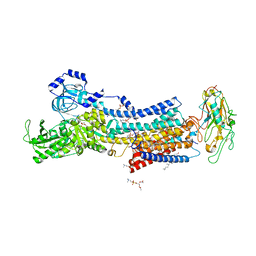 | | Crystal structures of Na+,K+-ATPase in complex with rostafuroxin | | Descriptor: | (3S,5R,8R,9S,10S,13S,14S,17S)-17-(furan-3-yl)-10,13-dimethyl-2,3,4,5,6,7,8,9,11,12,15,16-dodecahydro-1H-cyclopenta[a]phenanthrene-3,14,17-triol, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ogawa, H, Cornelius, F, Kanai, R, Motoyama, K, Vilsen, B, Toyoshima, C. | | Deposit date: | 2020-10-29 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Binding of cardiotonic steroids to Na + ,K + -ATPase in the E2P state.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8SVY
 
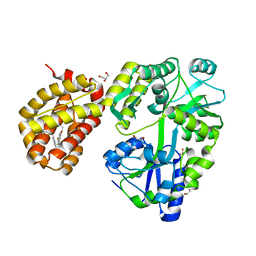 | | MBP-Mcl1 in complex with ligand 10 | | Descriptor: | (15P)-17-chloro-33-fluoro-12-[(2-methoxyethoxy)methyl]-5,14,22-trimethyl-28-oxa-9-thia-5,6,13,14,22-pentaazaheptacyclo[27.7.1.1~4,7~.0~11,15~.0~16,21~.0~20,24~.0~30,35~]octatriaconta-1(36),4(38),6,11(15),12,16,18,20,23,29(37),30,32,34-tridecaene-23-carboxylic acid, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Miller, B.R, Shaffer, P. | | Deposit date: | 2023-05-17 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Macrocyclic Carbon-Linked Pyrazoles As Novel Inhibitors of MCL-1.
Acs Med.Chem.Lett., 14, 2023
|
|
7DMQ
 
 | | Cryo-EM structure of LshCas13a-crRNA-anti-tag RNA complex | | Descriptor: | Anti-tag target RNA, CRISPR RNA, CRISPR/Cas system Cas13a | | Authors: | Wang, B, Zhang, T, Ding, J, Patel, D.J, Yang, H. | | Deposit date: | 2020-12-05 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis for self-cleavage prevention by tag:anti-tag pairing complementarity in type VI Cas13 CRISPR systems.
Mol.Cell, 81, 2021
|
|
