8R5V
 
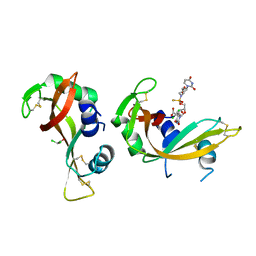 | | Crystal structure of bovine pancreatic ribonuclease A in complex with [Sp-PS]-mU-dT dinucleotide | | Descriptor: | 1-[(2~{R},4~{S},5~{R})-5-[[[(2~{R},6~{S})-2-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-6-(hydroxymethyl)morpholin-4-yl]-oxidanyl-phosphinothioyl]oxymethyl]-4-oxidanyl-oxolan-2-yl]-5-methyl-pyrimidine-2,4-dione, DICHLORO-ACETIC ACID, Ribonuclease pancreatic | | Authors: | Dolot, R, Jastrzebska, K, Antonczyk, P. | | Deposit date: | 2023-11-17 | | Release date: | 2023-12-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | P-Stereodefined morpholino dinucleoside 3',5'-phosphorothioates.
Org.Biomol.Chem., 22, 2024
|
|
4N93
 
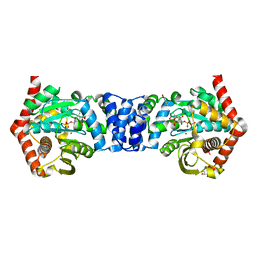 | | Alternative substrates of Mycobacterium tuberculosis anthranilate phosphoribosyl transferase | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 2-amino-6-methylbenzoic acid, Anthranilate phosphoribosyltransferase, ... | | Authors: | Castell, A, Short, F.L, Lott, J.S. | | Deposit date: | 2013-10-19 | | Release date: | 2014-04-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Alternative substrates reveal catalytic cycle and key binding events in the reaction catalysed by anthranilate phosphoribosyltransferase from Mycobacterium tuberculosis.
Biochem.J., 461, 2014
|
|
4N9F
 
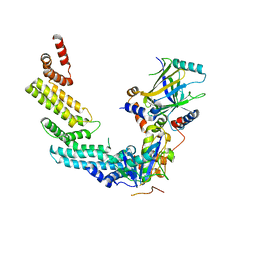 | | Crystal structure of the Vif-CBFbeta-CUL5-ElOB-ElOC pentameric complex | | Descriptor: | Core-binding factor subunit beta, Cullin-5, Transcription elongation factor B polypeptide 1, ... | | Authors: | Guo, Y.Y, Dong, L.Y, Huang, Z.W. | | Deposit date: | 2013-10-21 | | Release date: | 2014-01-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for hijacking CBF-b and CUL5 E3 ligase complex by HIV-1 Vif
Nature, 505, 2014
|
|
3HJO
 
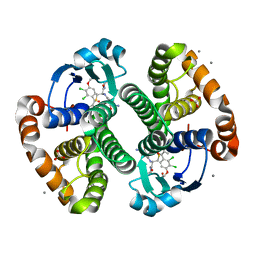 | |
3HJM
 
 | | Crystal structure of human Glutathione Transferase Pi Y108V mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CARBONATE ION, ... | | Authors: | Parker, L.J. | | Deposit date: | 2009-05-22 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Influence of the H-site residue 108 on human glutathione transferase P1-1 ligand binding: structure-thermodynamic relationships and thermal stability.
Protein Sci., 18, 2009
|
|
3HI6
 
 | |
3U23
 
 | | Atomic resolution crystal structure of the 2nd SH3 domain from human CD2AP (CMS) in complex with a proline-rich peptide from human RIN3 | | Descriptor: | 1,2-ETHANEDIOL, CD2-associated protein, Ras and Rab interactor 3 | | Authors: | Simister, P.C, Rouka, E, Janning, M, Muniz, J.R.C, Kirsch, K.H, Knapp, S, von Delft, F, Filippakopoulos, P, Arrowsmith, C.H, Krojer, T, Edwards, A.M, Weigelt, J, Bountra, C, Feller, S.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-09-30 | | Release date: | 2011-12-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Differential Recognition Preferences of the Three Src Homology 3 (SH3) Domains from the Adaptor CD2-associated Protein (CD2AP) and Direct Association with Ras and Rab Interactor 3 (RIN3).
J.Biol.Chem., 290, 2015
|
|
6DZ5
 
 | | Citrobacter freundii tyrosine phenol-lyase F448A mutant complexed with L-alanine | | Descriptor: | (2E)-3-(3-fluoro-4-hydroxyphenyl)-2-{[(Z)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4(1H)-ylidene}methyl]imino}propanoic acid, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, POTASSIUM ION, ... | | Authors: | Phillips, R.S. | | Deposit date: | 2018-07-03 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal Structures of Wild-Type and F448A Mutant Citrobacter freundii Tyrosine Phenol-Lyase Complexed with a Substrate and Inhibitors: Implications for the Reaction Mechanism.
Biochemistry, 57, 2018
|
|
6NQ4
 
 | |
2NQ0
 
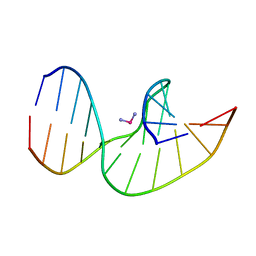 | | Solution Structures of a DNA Dodecamer Duplex with a Cisplatin 1,2-d(GG) Intrastrand Cross-Link | | Descriptor: | 5'-D(*CP*CP*TP*CP*AP*GP*GP*CP*CP*TP*CP*C)-3', 5'-D(*GP*GP*AP*GP*GP*CP*CP*TP*GP*AP*GP*G)-3', Cisplatin | | Authors: | Wu, Y, Bhattacharyya, D, Chaney, S, Campbell, S. | | Deposit date: | 2006-10-30 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of a DNA Dodecamer Duplex with and without a Cisplatin 1,2-d(GG) Intrastrand Cross-Link: Comparison with the Same DNA Duplex Containing an Oxaliplatin 1,2-d(GG) Intrastrand Cross-Link
Biochemistry, 46, 2007
|
|
4EGE
 
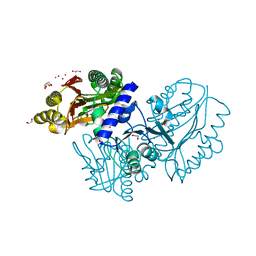 | |
4Y2T
 
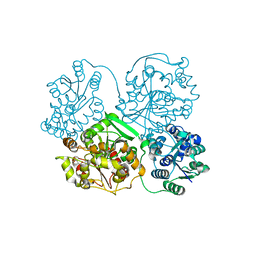 | |
7ONR
 
 | | PARP1 catalytic domain in complex with 8-chloroquinazolinone-based inhibitor (compound 9) | | Descriptor: | 8-chloranyl-2-[3-[4-(1,5-dimethylimidazol-2-yl)piperazin-1-yl]propyl]-3~{H}-quinazolin-4-one, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Schimpl, M, Balazs, A, Barratt, D, Bista, M, Chuba, M, Degorce, S.L, Di Fruscia, P, Embrey, K, Ghosh, A, Gill, S, Gunnarsson, A, Hande, S, Hemsley, P, Illuzzi, G, Lane, J, Larner, C, Leo, E, Madin, A, Martin, S, McWilliams, L, Orme, J, Pachl, F, Packer, M, Pike, A, Staniszewska, A.D, Talbot, V, Underwood, E, Varnes, G.J, Zhang, A, Zheng, X, Johannes, J.W. | | Deposit date: | 2021-05-25 | | Release date: | 2021-09-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of 5-{4-[(7-Ethyl-6-oxo-5,6-dihydro-1,5-naphthyridin-3-yl)methyl]piperazin-1-yl}- N -methylpyridine-2-carboxamide (AZD5305): A PARP1-DNA Trapper with High Selectivity for PARP1 over PARP2 and Other PARPs.
J.Med.Chem., 64, 2021
|
|
3GPJ
 
 | | Crystal structure of the yeast 20S proteasome in complex with syringolin B | | Descriptor: | N-{[(1S)-2-methyl-1-{[(5S,8S)-5-(1-methylethyl)-2,7-dioxo-1,6-diazacyclododec-3-en-8-yl]carbamoyl}propyl]carbamoyl}-L-valine, Proteasome component C1, Proteasome component C11, ... | | Authors: | Groll, M, Huber, R, Kaiser, M. | | Deposit date: | 2009-03-23 | | Release date: | 2009-06-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Synthetic and structural studies on syringolin A and B reveal critical determinants of selectivity and potency of proteasome inhibition
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1KQZ
 
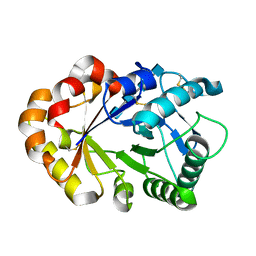 | | Hevamine Mutant D125A/E127A/Y183F in Complex with Tetra-NAG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hevamine A | | Authors: | Rozeboom, H.J, Dijkstra, B.W. | | Deposit date: | 2002-01-08 | | Release date: | 2002-01-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Expression and Characterization of Active Site Mutants of Hevamine, a
Chitinase from the Rubber Tree Hevea brasiliensis.
Eur.J.Biochem., 269, 2002
|
|
7O3H
 
 | | Murine CIII2 focus-refined from supercomplex CICIII2 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CARDIOLIPIN, ... | | Authors: | Vercellino, I, Sazanov, L.A. | | Deposit date: | 2021-04-01 | | Release date: | 2021-10-13 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure and assembly of the mammalian mitochondrial supercomplex CIII 2 CIV.
Nature, 598, 2021
|
|
9COP
 
 | | Yeast RAVE bound to V-ATPase V1 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Regulator of V-ATPase in vacuolar membrane protein 1, ... | | Authors: | Wang, H, Rubinstein, J.L. | | Deposit date: | 2024-07-17 | | Release date: | 2024-11-20 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure of yeast RAVE bound to a partial V 1 complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6ZWE
 
 | | Crystal structure of human acetylcholinesterase in complex with ((6-((2E,4E)-5-(benzo[d][1,3]dioxol-5-yl)penta-2,4-dienamido)hexyl)triphenylphosphonium bromide) | | Descriptor: | (2~{E},4~{E})-5-(1,3-benzodioxol-5-yl)-~{N}-[6-(triphenyl-$l^{5}-phosphanyl)hexyl]penta-2,4-dienamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Da Silva, O, Dias, J, Nachon, F, Brazzolotto, X. | | Deposit date: | 2020-07-28 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Fine-Tuning the Biological Profile of Multitarget Mitochondriotropic Antioxidants for Neurodegenerative Diseases.
Antioxidants (Basel), 10, 2021
|
|
9L3S
 
 | | Staphylococcus aureus lipase-Penfluridol complex (in space) | | Descriptor: | 1-[4,4-bis(4-fluorophenyl)butyl]-4-[4-chloranyl-3-(trifluoromethyl)phenyl]piperidin-4-ol, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kitadokoro, J, Hirokawa, T, Kamo, M, Furubayasi, N, Inaka, K, Kamitani, S, Kitadokoro, K. | | Deposit date: | 2024-12-19 | | Release date: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural analysis shows the mode of inhibition for Staphylococcus aureus lipase by antipsychotic penfluridol.
Sci Rep, 15, 2025
|
|
3TV5
 
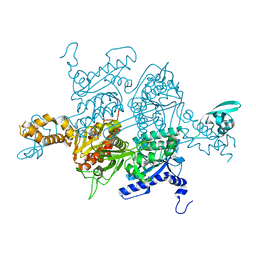 | | Crystal Structure of the humanized carboxyltransferase domain of yeast Acetyl-coA caroxylase in complex with compound 1 | | Descriptor: | (3R)-1'-(9-ANTHRYLCARBONYL)-3-(MORPHOLIN-4-YLCARBONYL)-1,4'-BIPIPERIDINE, Acetyl-CoA carboxylase | | Authors: | Rajamohan, F, Marr, E, Reyes, A, Landro, J.A, Anderson, M.D, Corbett, J.W, Dirico, K.J, Harwood, J.H, Tu, M, Vajdos, F.F. | | Deposit date: | 2011-09-19 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-guided Inhibitor Design for Human Acetyl-coenzyme A Carboxylase by Interspecies Active Site Conversion.
J.Biol.Chem., 286, 2011
|
|
8H97
 
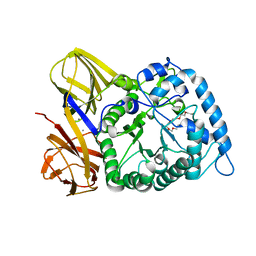 | | GH86 agarase Aga86A_Wa | | Descriptor: | Beta-agarase, CALCIUM ION, HEXAETHYLENE GLYCOL | | Authors: | Zhang, Y.Y, Dong, S, Feng, Y.G, Chang, Y.G. | | Deposit date: | 2022-10-25 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Structural characterization on a beta-agarase Aga86A_Wa from Wenyingzhuangia aestuarii reveals the prevalent methyl-galactose accommodation capacity of GH86 enzymes at subsite -1.
Carbohydr Polym, 306, 2023
|
|
7OET
 
 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH 1,5-dimethyl-N-(2-(methylamino)-2-oxo-1-(tetrahydro-2H-pyran-4-yl)ethyl)-6-oxo-N-(2-phenyl-2-(pyridin-2-yl)ethyl)-1,6-dihydropyridine-3-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 1,5-dimethyl-~{N}-[(1~{R})-2-(methylamino)-1-(oxan-4-yl)-2-oxidanylidene-ethyl]-6-oxidanylidene-~{N}-[(2~{S})-2-phenyl-2-pyridin-2-yl-ethyl]pyridine-3-carboxamide, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ... | | Authors: | Chung, C. | | Deposit date: | 2021-09-21 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Discovery of a Highly Selective BET BD2 Inhibitor from a DNA-Encoded Library Technology Screening Hit.
J.Med.Chem., 64, 2021
|
|
9L3C
 
 | | Staphylococcus aureus lipase-Penfluridol complex (on the ground) | | Descriptor: | 1-[4,4-bis(4-fluorophenyl)butyl]-4-[4-chloranyl-3-(trifluoromethyl)phenyl]piperidin-4-ol, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kitadokoro, J, Hirokawa, T, Kamitani, S, Kitadokoro, K. | | Deposit date: | 2024-12-18 | | Release date: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural analysis shows the mode of inhibition for Staphylococcus aureus lipase by antipsychotic penfluridol.
Sci Rep, 15, 2025
|
|
4Y1D
 
 | |
4Y2V
 
 | |
