6KVG
 
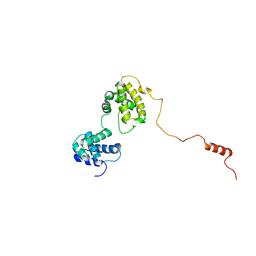 | | The solution structure of human Orc6 | | Descriptor: | Origin recognition complex subunit 6 | | Authors: | Liu, C, Xu, N, You, Y, Zhu, G. | | Deposit date: | 2019-09-04 | | Release date: | 2020-09-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of DNA replication origin recognition by human Orc6 protein binding with DNA.
Nucleic Acids Res., 48, 2020
|
|
8U1N
 
 | |
8TLT
 
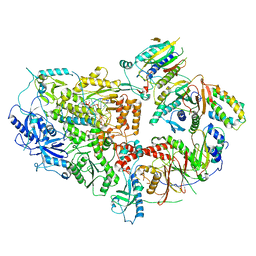 | | Cryo-EM structure of Rev1(deltaN)-Polzeta-DNA-dCTP complex | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*TP*AP*AP*TP*GP*GP*TP*AP*GP*GP*GP*GP*AP*GP*GP*GP*AP*AP*T)-3'), ... | | Authors: | Malik, R, Aggarwal, A.K. | | Deposit date: | 2023-07-27 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Cryo-EM structure of the Rev1-Pol zeta holocomplex reveals the mechanism of their cooperativity in translesion DNA synthesis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8TLQ
 
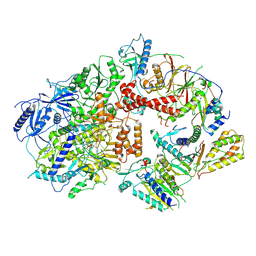 | | Cryo-EM structure of the Rev1-Polzeta-DNA-dCTP complex | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (30-MER), ... | | Authors: | Malik, R, Aggarwal, A.K. | | Deposit date: | 2023-07-27 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Cryo-EM structure of the Rev1-Pol zeta holocomplex reveals the mechanism of their cooperativity in translesion DNA synthesis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8TPQ
 
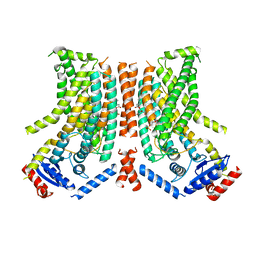 | |
8TOK
 
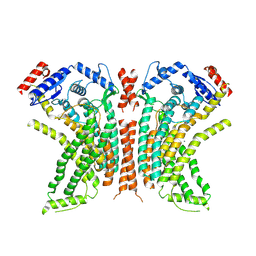 | |
8TPR
 
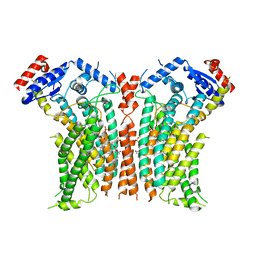 | |
8TPM
 
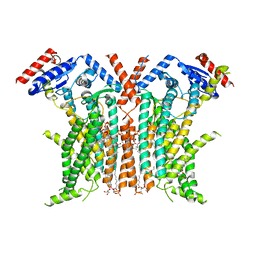 | |
8TPS
 
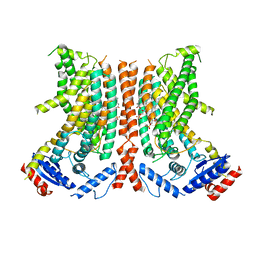 | |
8TPN
 
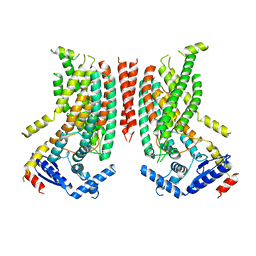 | |
8TOL
 
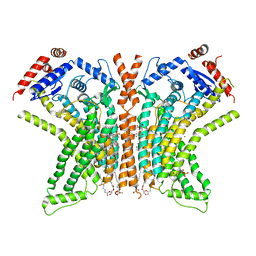 | |
8TOI
 
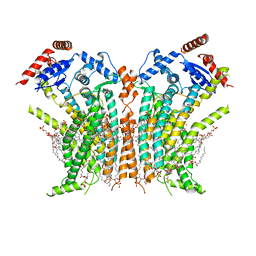 | |
8TPP
 
 | |
8TPT
 
 | |
8TNJ
 
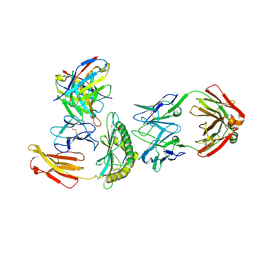 | | Cryo-EM structure of HLA-B*73:01 bound to a 9mer peptide and two Fabs | | Descriptor: | 9mer peptide,Beta-2-microglobulin,MHC class I antigen chimera, B.1 Fab heavy chain, B.1 Fab light chain, ... | | Authors: | Ross, P, Adams, E.J, Lodwick, J, Zhao, M, Slezak, T, Kossiakoff, A. | | Deposit date: | 2023-08-02 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of HLA-B*73:01 bound to a 9mer peptide and two Fabs
To Be Published
|
|
7Y1Q
 
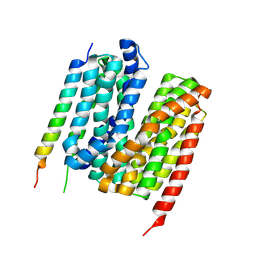 | | 5.0 angstrom cryo-EM structure of transmembrane regions of mouse Basigin/MCT1 in complex with antibody 6E7F1 | | Descriptor: | Isoform 2 of Basigin, Monocarboxylate transporter 1 | | Authors: | Zhang, H, Yang, X, Xue, Y, Huang, Y, Mo, X, Zhang, H, Li, N, Gao, N, Li, X, Wang, S, Gao, Y, Liao, J. | | Deposit date: | 2022-06-08 | | Release date: | 2023-06-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (5.03 Å) | | Cite: | Allosteric modulation of monocarboxylate transporters 1 and 4 by targeting their chaperon Basigin-2
To Be Published
|
|
8TKQ
 
 | |
8U5V
 
 | |
8U5X
 
 | |
8U1H
 
 | | Axle-less Bacillus sp. PS3 F1 ATPase mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase gamma chain, ATP synthase subunit alpha, ... | | Authors: | Furlong, E.J, Zeng, Y.C, Brown, S.H.J, Sobti, M, Stewart, A.G. | | Deposit date: | 2023-09-01 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The molecular structure of an axle-less F1 ATPase
To Be Published
|
|
8UDG
 
 | | S1V2-72 Fab bound to EHA2 from influenza B/Malaysia/2506/2004 | | Descriptor: | Hemagglutinin, S1V2-72 heavy chain, S1V2-72 light chain | | Authors: | Finney, J, Kong, S, Walsh Jr, R.M, Harrison, S.C, Kelsoe, G. | | Deposit date: | 2023-09-28 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (4.98 Å) | | Cite: | Protective human antibodies against a conserved epitope in pre- and postfusion influenza hemagglutinin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UXL
 
 | |
8UX1
 
 | |
8UXM
 
 | |
8UR3
 
 | |
