8CH6
 
 | | Structure of a late-stage activated spliceosome (BAqr) arrested with a dominant-negative Aquarius mutant (state B complex). | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Cell division cycle 5-like protein, ... | | Authors: | Cretu, C, Schmitzova, J, Pena, V. | | Deposit date: | 2023-02-07 | | Release date: | 2023-05-10 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural basis of catalytic activation in human splicing.
Nature, 617, 2023
|
|
8CGN
 
 | | Non-rotated 80S S. cerevisiae ribosome with ligands | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Milicevic, N, Jenner, L, Myasnikov, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2023-02-06 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.28 Å) | | Cite: | mRNA reading frame maintenance during eukaryotic ribosome translocation.
Nature, 625, 2024
|
|
8CG8
 
 | | Translocation intermediate 3 (TI-3) of 80S S. cerevisiae ribosome with ligands and eEF2 in the presence of sordarin | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Milicevic, N, Jenner, L, Myasnikov, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2023-02-03 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | mRNA reading frame maintenance during eukaryotic ribosome translocation.
Nature, 625, 2024
|
|
8CF5
 
 | | Translocation intermediate 1 (TI-1) of 80S S. cerevisiae ribosome with ligands and eEF2 in the presence of sordarin | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Milicevic, N, Jenner, L, Myasnikov, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2023-02-02 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | mRNA reading frame maintenance during eukaryotic ribosome translocation.
Nature, 625, 2024
|
|
8G1Q
 
 | | Co-crystal structure of Compound 1 in complex with the bromodomain of human SMARCA4 and pVHL:ElonginC:ElonginB | | Descriptor: | DI(HYDROXYETHYL)ETHER, Elongin-B, Elongin-C, ... | | Authors: | Ghimire Rijal, S, Wurz, R.P, Vaish, A. | | Deposit date: | 2023-02-02 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.73 Å) | | Cite: | Affinity and cooperativity modulate ternary complex formation to drive targeted protein degradation.
Nat Commun, 14, 2023
|
|
8G1P
 
 | | Co-crystal structure of Compound 11 in complex with the bromodomain of human SMARCA2 and pVHL:ElonginC:ElonginB | | Descriptor: | (2~{S},4~{R})-~{N}-[[2-[2-[4-[[4-[3-azanyl-6-(2-hydroxyphenyl)pyridazin-4-yl]piperazin-1-yl]methyl]phenyl]ethoxy]-4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-1-[(2~{S})-2-[(1-fluoranylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Ghimire Rijal, S, Wurz, R.P, Vaish, A. | | Deposit date: | 2023-02-02 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Affinity and cooperativity modulate ternary complex formation to drive targeted protein degradation.
Nat Commun, 14, 2023
|
|
8CEH
 
 | | Translocation intermediate 4 (TI-4) of 80S S. cerevisiae ribosome with ligands and eEF2 in the presence of sordarin | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Milicevic, N, Jenner, L, Myasnikov, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2023-02-01 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.05 Å) | | Cite: | mRNA reading frame maintenance during eukaryotic ribosome translocation.
Nature, 625, 2024
|
|
8I7J
 
 | |
8CDR
 
 | | Translocation intermediate 2 (TI-2) of 80S S. cerevisiae ribosome with ligands and eEF2 in the presence of sordarin | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Milicevic, N, Jenner, L, Myasnikov, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2023-01-31 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.04 Å) | | Cite: | mRNA reading frame maintenance during eukaryotic ribosome translocation.
Nature, 625, 2024
|
|
8CDL
 
 | | 80S S. cerevisiae ribosome with ligands in hybrid-2 pre-translocation (PRE-H2) complex | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Milicevic, N, Jenner, L, Myasnikov, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2023-01-31 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | mRNA reading frame maintenance during eukaryotic ribosome translocation.
Nature, 625, 2024
|
|
8CCS
 
 | | 80S S. cerevisiae ribosome with ligands in hybrid-1 pre-translocation (PRE-H1) complex | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Milicevic, N, Jenner, L, Myasnikov, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2023-01-27 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (1.97 Å) | | Cite: | mRNA reading frame maintenance during eukaryotic ribosome translocation.
Nature, 625, 2024
|
|
8FY0
 
 | | E3:PROTAC:target ternary complex structure (VCB/753b/BCL-xL) | | Descriptor: | Bcl-2-like protein 1, CACODYLIC ACID, Elongin-B, ... | | Authors: | Olsen, S.K, Nayak, D, Lv, D, Yuan, Y, Zhang, P, Hu, W, Lv, Z, Sung, P, Hromas, R, Zheng, G, Zhou, D. | | Deposit date: | 2023-01-25 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Development and crystal structures of a potent second-generation dual degrader of BCL-2 and BCL-xL.
Nat Commun, 15, 2024
|
|
8FY2
 
 | | E3:PROTAC:target ternary complex structure (VCB/WH244/BCL-2) | | Descriptor: | Apoptosis regulator Bcl-2, Elongin-B, Elongin-C, ... | | Authors: | Nayak, D, Lv, D, Yuan, Y, Zhang, P, Hu, W, Ruben, E, Lv, Z, Sung, P, Hromas, R, Zheng, G, Zhou, D, Olsen, S.K. | | Deposit date: | 2023-01-25 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Development and crystal structures of a potent second-generation dual degrader of BCL-2 and BCL-xL.
Nat Commun, 15, 2024
|
|
8FY1
 
 | | E3:PROTAC:target ternary complex structure (VCB/753b/BCL-2) | | Descriptor: | Apoptosis regulator Bcl-2, Elongin-B, Elongin-C, ... | | Authors: | Nayak, D, Lv, D, Yuan, Y, Zhang, P, Hu, W, Lv, Z, Sung, P, Hromas, R, Zheng, G, Zhou, D, Olsen, S.K. | | Deposit date: | 2023-01-25 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Development and crystal structures of a potent second-generation dual degrader of BCL-2 and BCL-xL.
Nat Commun, 15, 2024
|
|
8CAS
 
 | | Cryo-EM structure of native Otu2-bound ubiquitinated 48S initiation complex (partial) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Ikeuchi, K, Buschauer, R, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2023-01-24 | | Release date: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for recognition and deubiquitination of 40S ribosomes by Otu2.
Nat Commun, 14, 2023
|
|
8CAH
 
 | | Cryo-EM structure of native Otu2-bound ubiquitinated 43S pre-initiation complex | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Ikeuchi, K, Buschauer, R, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2023-01-24 | | Release date: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis for recognition and deubiquitination of 40S ribosomes by Otu2.
Nat Commun, 14, 2023
|
|
8CAF
 
 | | N8C_Fab3b in complex with NEDD8-CUL1(WHB) | | Descriptor: | Cullin-1, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Duda, D.M, Yanishevski, D, Henneberg, L.T, Schulman, B.A. | | Deposit date: | 2023-01-24 | | Release date: | 2023-09-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Activity-based profiling of cullin-RING E3 networks by conformation-specific probes.
Nat.Chem.Biol., 19, 2023
|
|
8FVJ
 
 | | Dimeric form of HIV-1 Vif in complex with human CBF-beta, ELOB, ELOC, and CUL5 | | Descriptor: | Core-binding factor subunit beta, Cullin-5, Elongin-B, ... | | Authors: | Ito, F, Alvarez-Cabrera, A.L, Zhou, Z.H, Chen, X.S. | | Deposit date: | 2023-01-19 | | Release date: | 2023-09-06 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural basis of HIV-1 Vif-mediated E3 ligase targeting of host APOBEC3H.
Nat Commun, 14, 2023
|
|
8FVI
 
 | | Human APOBEC3H bound to HIV-1 Vif in complex with CBF-beta, ELOB, ELOC, and CUL5 | | Descriptor: | Core-binding factor subunit beta, Cullin 5, DNA dC->dU-editing enzyme APOBEC-3H, ... | | Authors: | Ito, F, Alvarez-Cabrera, A.L, Zhou, Z.H, Chen, X.S. | | Deposit date: | 2023-01-19 | | Release date: | 2023-09-06 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structural basis of HIV-1 Vif-mediated E3 ligase targeting of host APOBEC3H.
Nat Commun, 14, 2023
|
|
8C83
 
 | | Cryo-EM structure of in vitro reconstituted Otu2-bound Ub-40S complex | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S11-A, ... | | Authors: | Ikeuchi, K, Buschauer, R, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2023-01-18 | | Release date: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis for recognition and deubiquitination of 40S ribosomes by Otu2.
Nat Commun, 14, 2023
|
|
8FTQ
 
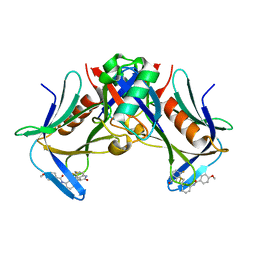 | | Crystal structure of hRpn13 Pru domain in complex with Ubiquitin and XL44 | | Descriptor: | N-(3-{[(3R)-5-fluoro-2-oxo-2,3-dihydro-1H-indol-3-yl]methyl}phenyl)-4-methoxybenzamide, Proteasomal ubiquitin receptor ADRM1, Ubiquitin | | Authors: | Walters, K.J, Lu, X, Chandravanshi, M. | | Deposit date: | 2023-01-13 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structure-based designed small molecule depletes hRpn13 Pru and a select group of KEN box proteins.
Nat Commun, 15, 2024
|
|
8FRU
 
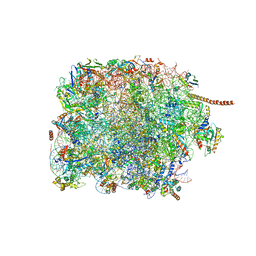 | | 60S subunit of the Giardia lamblia 80S ribosome | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Eiler, D.R, Wimberly, B.T, Bilodeau, D.Y, Rissland, O.S, Kieft, J.S. | | Deposit date: | 2023-01-08 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | The Giardia lamblia ribosome structure reveals divergence in several biological pathways and the mode of emetine function.
Structure, 32, 2024
|
|
8C3A
 
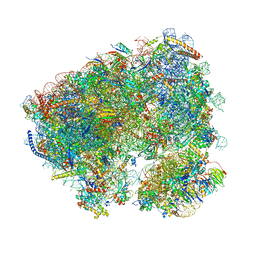 | | Crystal structure of ailanthone bound to the Candida albicans 80S ribosome | | Descriptor: | 18S, 25S, 3-O-acetyl-2-O-(3-O-acetyl-6-deoxy-beta-D-glucopyranosyl)-6-deoxy-1-O-{[(2R,2'S,3a'R,4''S,5''R,6'S,7a'S)-5''-methyl-4''-{[(2E)-3-phenylprop-2-enoyl]oxy}decahydrodispiro[oxirane-2,3'-[1]benzofuran-2',2''-pyran]-6'-yl]carbonyl}-beta-D-glucopyranose, ... | | Authors: | Kolosova, O, Zgadzay, Y, Yusupov, M. | | Deposit date: | 2022-12-23 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | New crystal system to promote screening for new eukaryotic inhibitors
To Be Published
|
|
8HTD
 
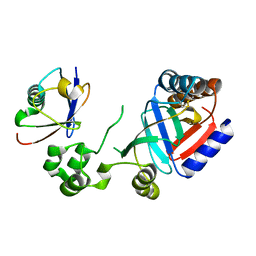 | | Crystal structure of an effector from Chromobacterium violaceum in complex with ubiquitin | | Descriptor: | NAD(+)--protein-threonine ADP-ribosyltransferase, Ubiquitin | | Authors: | Tan, J, Wang, X, Zhou, Y, Zhu, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-22 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Molecular basis of threonine ADP-ribosylation of ubiquitin by bacterial ARTs.
Nat.Chem.Biol., 20, 2024
|
|
8HTE
 
 | | Crystal structure of an effector mutant in complex with ubiquitin | | Descriptor: | GLYCEROL, NAD(+)--protein-threonine ADP-ribosyltransferase, NICOTINAMIDE, ... | | Authors: | Tan, J, Wang, X, Zhou, Y, Zhu, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-22 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.307 Å) | | Cite: | Molecular basis of threonine ADP-ribosylation of ubiquitin by bacterial ARTs.
Nat.Chem.Biol., 20, 2024
|
|
