7AEN
 
 | | Galectin-8 N-terminal carbohydrate recognition domain in complex with methyl 3-O-((7-carboxy)quinolin-2-yl)-methoxy)-beta-D-galactopyranoside | | Descriptor: | CHLORIDE ION, GLYCEROL, Isoform 2 of Galectin-8, ... | | Authors: | Hassan, M, Klavern, V.S, Hakansson, M, Anderluh, M, Tomasic, T, Jakopin, Z, Nilsson, J.U, Kovacic, R, Walse, B, Diehl, C. | | Deposit date: | 2020-09-17 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Design of d-Galactal Derivatives with High Affinity and Selectivity for the Galectin-8 N-Terminal Domain
Acs Med.Chem.Lett., 12, 2021
|
|
8DG6
 
 | |
8Y8R
 
 | | Cryo-EM structure of AQP3 Y212T in DDM micelle | | Descriptor: | Aquaporin-3, DODECYL-BETA-D-MALTOSIDE | | Authors: | Kozai, D, Suzuki, S, Kamegawa, A, Nishikawa, K, Suzuki, H, Fujiyoshi, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2025-03-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Narrowed pore conformations of aquaglyceroporins AQP3 and GlpF.
Nat Commun, 16, 2025
|
|
8VZ9
 
 | | Crystal structure of mouse MAIT M2A TCR-MR1-5-OP-RU complex | | Descriptor: | 1-deoxy-1-({2,6-dioxo-5-[(E)-propylideneamino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Ciacchi, L, Rossjohn, J, Awad, W. | | Deposit date: | 2024-02-11 | | Release date: | 2024-04-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Mouse mucosal-associated invariant T cell receptor recognition of MR1 presenting the vitamin B metabolite, 5-(2-oxopropylideneamino)-6-d-ribitylaminouracil.
J.Biol.Chem., 300, 2024
|
|
6OJV
 
 | | Crystal structure of human thymidylate synthase delta(7-29) in complex with dUMP and 2-amino-4-oxo-4,7-dihydro-pyrrolo[2,3-d]pyrimidine-methyl-phenyl-L-glutamic acid | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, N-{4-[(2-amino-4-hydroxy-7H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl]benzoyl}-L-glutamic acid, Thymidylate synthase,Thymidylate synthase | | Authors: | Czyzyk, D.J, Anderson, K.S, Valhondo, M, Jorgensen, W.L. | | Deposit date: | 2019-04-12 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.593 Å) | | Cite: | Understanding the structural basis of species selective, stereospecific inhibition for Cryptosporidium and human thymidylate synthase.
Febs Lett., 593, 2019
|
|
1EEO
 
 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B COMPLEXED WITH ACETYL-E-L-E-F-PTYR-M-D-Y-E-NH2 | | Descriptor: | ACETYL-E-L-E-F-PTYR-M-D-Y-E-NH2 PEPTIDE, MAGNESIUM ION, PROTEIN TYROSINE PHOSPHATASE 1B | | Authors: | Sarmiento, M, Puius, Y.A, Vetter, S.W, Lawrence, D.S, Almo, S.C, Zhang, Z.Y. | | Deposit date: | 2000-02-01 | | Release date: | 2001-02-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of plasticity in protein tyrosine phosphatase 1B substrate recognition.
Biochemistry, 39, 2000
|
|
1E55
 
 | | Crystal structure of the inactive mutant Monocot (Maize ZMGlu1) beta-glucosidase ZMGluE191D in complex with the competitive inhibitor dhurrin | | Descriptor: | (2S)-HYDROXY(4-HYDROXYPHENYL)ETHANENITRILE, BETA-GLUCOSIDASE, beta-D-glucopyranose | | Authors: | Czjzek, M, Cicek, M, Bevan, D.R, Zamboni, V, Henrissat, B, Esen, A. | | Deposit date: | 2000-07-18 | | Release date: | 2000-12-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The mechanism of substrate (aglycone) specificity in beta-glucosidases is revealed by crystal structures of mutant maize beta-glucosidase-DIMBOA, -DIMBOAGlc, and -dhurrin complexes.
Proc. Natl. Acad. Sci. U.S.A., 97, 2000
|
|
1E7P
 
 | | QUINOL:FUMARATE REDUCTASE FROM WOLINELLA SUCCINOGENES | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Lancaster, C.R.D, Kroeger, A. | | Deposit date: | 2000-09-01 | | Release date: | 2001-04-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A Third Crystal Form of Wolinella Succinogenes Quinol:Fumarate Reductase Reveals Domain Closure at the Site of Fumarate Reduction
Eur.J.Biochem., 268, 2001
|
|
7Q3S
 
 | | Crystal structure of UGT706F8 from Zea mays | | Descriptor: | Glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Fredslund, F, Bidart, G.N, Welner, D.H. | | Deposit date: | 2021-10-28 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Family 1 Glycosyltransferase UGT706F8 from Zea mays Selectively Catalyzes the Synthesis of Silibinin 7- O -beta-d-Glucoside.
Acs Sustain Chem Eng, 10, 2022
|
|
6OJA
 
 | | Crystal structure of the N. meningitides methionine-binding protein in its L-methionine bound conformation | | Descriptor: | Lipoprotein, METHIONINE | | Authors: | Nguyen, P.T, Lai, J.Y, Kaiser, J.T, Rees, D.C. | | Deposit date: | 2019-04-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures of the Neisseria meningitides methionine-binding protein MetQ in substrate-free form and bound to l- and d-methionine isomers.
Protein Sci., 28, 2019
|
|
8B54
 
 | | CDK2/cyclin A2 in complex with pyrazolo[4,3-d]pyrimidine inhibitor LGR6768 | | Descriptor: | 1,2-ETHANEDIOL, Cyclin-A2, Cyclin-dependent kinase 2, ... | | Authors: | Djukic, S, Skerlova, J, Rezacova, P. | | Deposit date: | 2022-09-21 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Characterization of new highly selective pyrazolo[4,3-d]pyrimidine inhibitor of CDK7.
Biomed Pharmacother, 161, 2023
|
|
8WWW
 
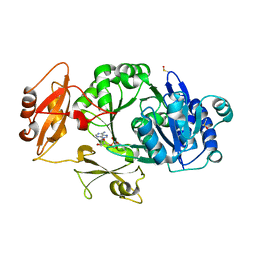 | | Crystal structure of the adenylation domain of SyAstC | | Descriptor: | Carrier domain-containing protein, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Li, Z, Zhu, Z, Zhu, D, Shen, Y. | | Deposit date: | 2023-10-27 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Functional Application of the Single-Module NRPS-like d-Alanyltransferase in Maytansinol Biosynthesis
Acs Catalysis, 14, 2024
|
|
7R2I
 
 | |
7R2L
 
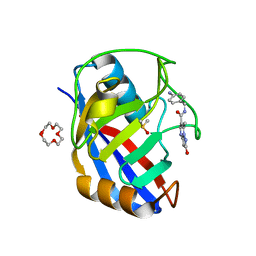 | | Human Cyclophilin D in complex with N-(4-aminophenyl)-7-methyl-2-oxo-1H,2H-pyrazolo[1,5-a]pyrimidine-6-carboxamide | | Descriptor: | DIMETHYL SULFOXIDE, N-(4-aminophenyl)-7-methyl-2-oxo-1H,2H-pyrazolo[1,5-a]pyrimidine-6-carboxamide, Peptidyl-prolyl cis-trans isomerase F, ... | | Authors: | Silva, D.O, Graedler, U, Bandeiras, T.M. | | Deposit date: | 2022-02-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Human Cyclophilin D in complex with
N-(4-aminophenyl)-7-methyl-2-oxo-1H,2H-pyrazolo[1,5-a]pyrimidine-6-carboxamide
To Be Published
|
|
7R2J
 
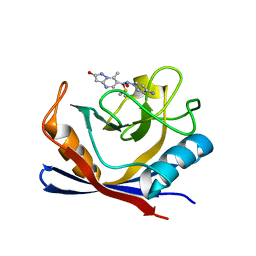 | | Human Cyclophilin D in complex with N-(4-aminophenyl)-7-methyl-2-oxo-1H,2H-pyrazolo[1,5-a]pyrimidine-6-carboxamide | | Descriptor: | DIMETHYL SULFOXIDE, N-(4-aminophenyl)-7-methyl-2-oxo-1H,2H-pyrazolo[1,5-a]pyrimidine-6-carboxamide, Peptidyl-prolyl cis-trans isomerase F, ... | | Authors: | Silva, D.O, Graedler, U, Bandeiras, T.M. | | Deposit date: | 2022-02-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Human Cyclophilin D in complex with
N-(4-aminophenyl)-7-methyl-2-oxo-1H,2H-pyrazolo[1,5-a]pyrimidine-6-carboxamide
To Be Published
|
|
6N9O
 
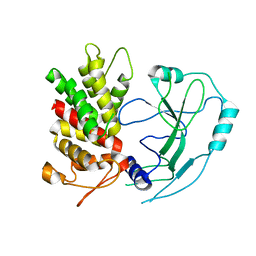 | | Crystal structure of human GSDMD | | Descriptor: | Gasdermin-D | | Authors: | Liu, Z, Wang, C, Yang, J, Xiao, T.S. | | Deposit date: | 2018-12-03 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal Structures of the Full-Length Murine and Human Gasdermin D Reveal Mechanisms of Autoinhibition, Lipid Binding, and Oligomerization.
Immunity, 51, 2019
|
|
5L7V
 
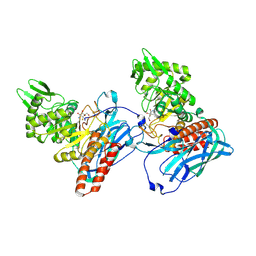 | | Crystal Structure of BvGH123 with bond transition state analog Galthiazoline. | | Descriptor: | (3aR,5R,6R,7R,7aR)-5-(hydroxymethyl)-2-methyl-5,6,7,7a-tetrahydro-3aH-pyrano[3,2-d][1,3]thiazole-6,7-diol, glycoside hydrolase | | Authors: | Roth, C, Petricevic, M, John, A, Goddard-Borger, E.D, Davies, G.J, Williams, S.J. | | Deposit date: | 2016-06-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and mechanistic insights into a Bacteroides vulgatus retaining N-acetyl-beta-galactosaminidase that uses neighbouring group participation.
Chem. Commun. (Camb.), 52, 2016
|
|
8DEA
 
 | |
7PMT
 
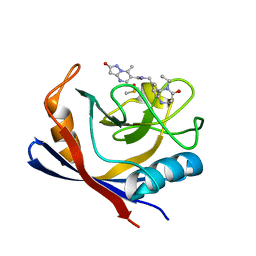 | | Human Cyclophilin D in complex with N-[(5-ethyl-4-oxo-1,2,3,4,5,6- hexahydro-1,5-benzodiazocin-8-yl)methyl]-7-methyl-2-oxo-1H,2H-pyrazolo[1,5-a]pyrimidine-6-carboxamide | | Descriptor: | DIMETHYL SULFOXIDE, Peptidyl-prolyl cis-trans isomerase F, mitochondrial, ... | | Authors: | Silva, D.O, Graedler, U, Bandeiras, T.M. | | Deposit date: | 2021-09-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Human Cyclophilin D in complex with N-[(4-aminophenyl)methyl]-7-methyl-2-oxo-1H,2H-pyrazolo[1,5-a]pyrimidine-6-carboxamide
To Be Published
|
|
8VZ8
 
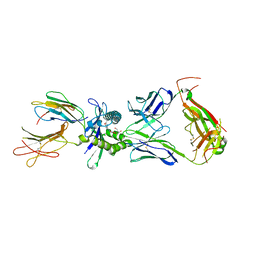 | | Crystal structure of mouse MAIT M2B TCR-MR1-5-OP-RU complex | | Descriptor: | 1-deoxy-1-({2,6-dioxo-5-[(E)-propylideneamino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Ciacchi, L, Rossjohn, J, Awad, W. | | Deposit date: | 2024-02-11 | | Release date: | 2024-04-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Mouse mucosal-associated invariant T cell receptor recognition of MR1 presenting the vitamin B metabolite, 5-(2-oxopropylideneamino)-6-d-ribitylaminouracil.
J.Biol.Chem., 300, 2024
|
|
8R2I
 
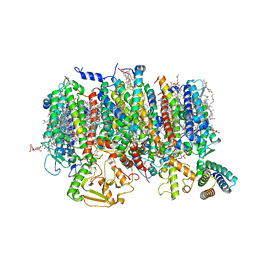 | | Cryo-EM Structure of native Photosystem II assembly intermediate from Chlamydomonas reinhardtii | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, ... | | Authors: | Fadeeva, M, Klaiman, D, Kandiah, E, Nelson, N. | | Deposit date: | 2023-11-06 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of native photosystem II assembly intermediate from Chlamydomonas reinhardtii .
Front Plant Sci, 14, 2023
|
|
6NZ8
 
 | | Structure of carbamylated apo OXA-231 carbapenemase | | Descriptor: | Beta-lactamase OXA-231, CHLORIDE ION, SODIUM ION | | Authors: | Favaro, D.C, Llontop, E.E, Vasconcelos, F.N, Antunes, V.U, Farah, S.C, Lincopan, N. | | Deposit date: | 2019-02-13 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Importance of the beta 5-beta 6 Loop for the Structure, Catalytic Efficiency, and Stability of Carbapenem-Hydrolyzing Class D beta-Lactamase Subfamily OXA-143.
Biochemistry, 58, 2019
|
|
6OME
 
 | |
6Y6F
 
 | | Crystal structure of STK17B (DRAK2) in complex with PKIS43 | | Descriptor: | 1,2-ETHANEDIOL, 2-[6-(4-methylsulfanylphenyl)thieno[3,2-d]pyrimidin-4-yl]sulfanylethanoic acid, Serine/threonine-protein kinase 17B | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Drewry, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-26 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | A Chemical Probe for Dark Kinase STK17B Derives Its Potency and High Selectivity through a Unique P-Loop Conformation.
J.Med.Chem., 63, 2020
|
|
6Y6H
 
 | | Crystal structure of STK17b (DRAK2) in complex with UNC-AP-194 probe | | Descriptor: | 1,2-ETHANEDIOL, 2-[6-(1-benzothiophen-2-yl)thieno[3,2-d]pyrimidin-4-yl]sulfanylethanoic acid, Serine/threonine-protein kinase 17B | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Drewry, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-26 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Chemical Probe for Dark Kinase STK17B Derives Its Potency and High Selectivity through a Unique P-Loop Conformation.
J.Med.Chem., 63, 2020
|
|
