6D0M
 
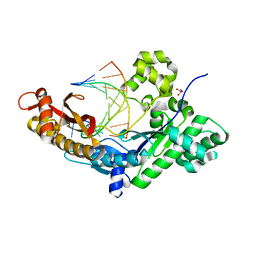 | | Polymerase Eta post-insertion binary complex with cytarabine (AraC) | | Descriptor: | DNA (5'-D(*CP*AP*TP*GP*AP*CP*AP*GP*TP*GP*CP*T)-3'), DNA polymerase eta, DNA/RNA (5'-D(*AP*GP*CP*AP*CP*TP*GP*T)-R(P*(CAR))-3'), ... | | Authors: | Rechkoblit, O, Aggarwal, A.K. | | Deposit date: | 2018-04-10 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.832 Å) | | Cite: | Structural basis for polymerase eta-promoted resistance to the anticancer nucleoside analog cytarabine.
Sci Rep, 8, 2018
|
|
7CRO
 
 | | NSD2 bearing E1099K/T1150A dual mutation in complex with 187-bp NCP | | Descriptor: | DNA (168-MER), Histone H2A, Histone H2B, ... | | Authors: | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | Deposit date: | 2020-08-14 | | Release date: | 2020-10-21 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
3NNN
 
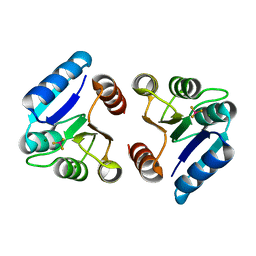 | | BeF3 Activated DrrD Receiver Domain | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, DNA BINDING RESPONSE REGULATOR D, MAGNESIUM ION | | Authors: | Robinson, V.L, Stock, A.M. | | Deposit date: | 2010-06-23 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Regulation of response regulator autophosphorylation through interdomain contacts.
J.Biol.Chem., 285, 2010
|
|
349D
 
 | |
5ZXM
 
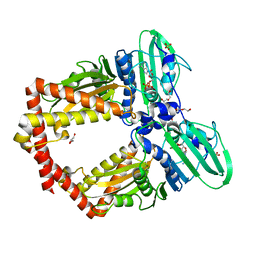 | | Crystal Structure of GyraseB N-terminal at 1.93A Resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, DNA gyrase subunit B, ... | | Authors: | Tiwari, P, Gupta, D, Sachdeva, E, Sharma, S, Singh, T.P, Ethayathulla, A.S, Kaur, P. | | Deposit date: | 2018-05-21 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.938 Å) | | Cite: | Structural insights into the transient closed conformation and pH dependent ATPase activity of S.Typhi GyraseB N- terminal domain.
Arch.Biochem.Biophys., 701, 2021
|
|
5LQG
 
 | |
7NL0
 
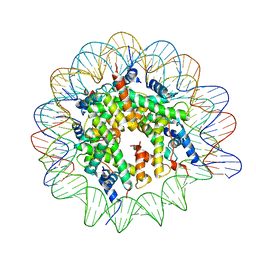 | | Cryo-EM structure of the Lin28B nucleosome core particle | | Descriptor: | DNA (131-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Roberts, G.A, Ozkan, B, Gachulincova, I, O Dwyer, M.R, Hall-Ponsele, E, Saxena, M, Robinson, P.J, Soufi, A. | | Deposit date: | 2021-02-19 | | Release date: | 2021-08-11 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Dissecting OCT4 defines the role of nucleosome binding in pluripotency.
Nat.Cell Biol., 23, 2021
|
|
6OM3
 
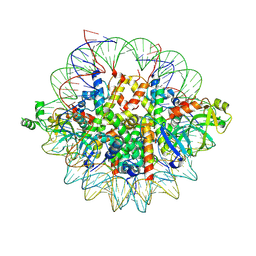 | | Crystal structure of the Orc1 BAH domain in complex with a nucleosome core particle | | Descriptor: | DNA (146-MER), DNA (147-MER), Histone H2A, ... | | Authors: | De Ioannes, P.E, Wang, M, Armache, K.-J. | | Deposit date: | 2019-04-18 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure and function of the Orc1 BAH-nucleosome complex.
Nat Commun, 10, 2019
|
|
5MVB
 
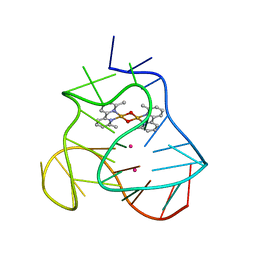 | | Solution structure of a human G-Quadruplex hybrid-2 form in complex with a Gold-ligand. | | Descriptor: | DNA (26-MER), POTASSIUM ION, bis(m2-Oxo)-bis(2-methyl-2,2'-bipyridine)-di-gold(iii) | | Authors: | Wirmer-Bartoschek, J, Jonker, H.R.A, Bendel, L.E, Gruen, T, Bazzicalupi, C, Messori, L, Gratteri, P, Schwalbe, H. | | Deposit date: | 2017-01-16 | | Release date: | 2017-05-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of a Ligand/Hybrid-2-G-Quadruplex Complex Reveals Rearrangements that Affect Ligand Binding.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
7K61
 
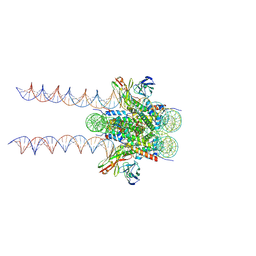 | | Cryo-EM structure of 197bp nucleosome aided by scFv | | Descriptor: | DNA (197-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Zhou, B.-R, Bai, Y. | | Deposit date: | 2020-09-17 | | Release date: | 2020-11-25 | | Last modified: | 2021-01-20 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Distinct Structures and Dynamics of Chromatosomes with Different Human Linker Histone Isoforms.
Mol.Cell, 81, 2021
|
|
8OF4
 
 | | Nucleosome Bound human SIRT6 (Composite) | | Descriptor: | DNA (145-MER), Histone H2A type 1, Histone H2B, ... | | Authors: | Smirnova, E, Bignon, E, Schultz, P, Papai, G, Ben-Shem, A. | | Deposit date: | 2023-03-13 | | Release date: | 2023-08-09 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Binding to nucleosome poises human SIRT6 for histone H3 deacetylation.
Elife, 12, 2024
|
|
5LQH
 
 | |
1F6V
 
 | |
1RYQ
 
 | | Putative DNA-directed RNA polymerase, subunit e'' from Pyrococcus Furiosus Pfu-263306-001 | | Descriptor: | DNA-directed RNA polymerase, subunit e'', ZINC ION | | Authors: | Liu, Z.-J, Chen, L, Tempel, W, Shah, A, Arendall III, W.B, Rose, J.P, Brereton, P.S, Izumi, M, Jenney Jr, F.E, Lee, H.S, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-12-22 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Parameter-space screening: a powerful tool for high-throughput crystal structure determination.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
5D4B
 
 | | Structural Basis for a New Templated Activity by Terminal Deoxynucleotidyl Transferase: Implications for V(D)J Recombination | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*C)-3'), DNA (5'-D(*TP*TP*TP*TP*TP*GP*G)-3'), MAGNESIUM ION, ... | | Authors: | Loc'h, J, Rosario, S, Delarue, M. | | Deposit date: | 2015-08-07 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structural Basis for a New Templated Activity by Terminal Deoxynucleotidyl Transferase: Implications for V(D)J Recombination.
Structure, 24, 2016
|
|
133D
 
 | | THE CRYSTAL STRUCTURE OF N4-METHYLCYTOSINE.GUANOSIN BASE-PAIRS IN THE SYNTHETIC HEXANUCLEOTIDE D(CGCGM(4)CG) | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*(C34)P*G)-3') | | Authors: | Cervi, A.R, Guy, A, Leonard, G.A, Teoule, R, Hunter, W.N. | | Deposit date: | 1993-07-29 | | Release date: | 1994-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of N4-methylcytosine.guanosine base-pairs in the synthetic hexanucleotide d(CGCGm4CG).
Nucleic Acids Res., 21, 1993
|
|
5Y27
 
 | | Crystal structure of Se-Met Dpb4-Dpb3 | | Descriptor: | DNA polymerase epsilon subunit D, GLYCEROL, Putative transcription factor C16C4.22 | | Authors: | Li, Y, Gao, F, Su, M, Zhang, F.B, Chen, Y.H. | | Deposit date: | 2017-07-24 | | Release date: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Coordinated regulation of heterochromatin inheritance by Dpb3-Dpb4 complex.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5Y26
 
 | | Crystal structure of native Dpb4-Dpb3 | | Descriptor: | DNA polymerase epsilon subunit D, GLYCEROL, Putative transcription factor C16C4.22 | | Authors: | Chen, Y.H, Li, Y, Gao, F. | | Deposit date: | 2017-07-24 | | Release date: | 2018-01-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Coordinated regulation of heterochromatin inheritance by Dpb3-Dpb4 complex.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4LOX
 
 | |
4XNO
 
 | | Crystal structure of 5'-CTTATPPPZZZATAAG | | Descriptor: | DNA (5'-D(*CP*TP*(BRU)P*AP*TP*(1WA)P*(1WA)P*(1WA)P*(1W5)P*(1W5)P*(1W5)P*AP*TP*AP*AP*G)-3') | | Authors: | Georgiadis, M.M, Singh, I. | | Deposit date: | 2015-01-15 | | Release date: | 2015-05-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.985 Å) | | Cite: | Structural basis for a six nucleotide genetic alphabet.
J. Am. Chem. Soc., 137, 2015
|
|
8J1J
 
 | | Cryo-EM structure of the AsCas12f-YHAM-sgRNAS3-5v7-target DNA | | Descriptor: | DNA (38-MER), MAGNESIUM ION, RNA (118-MER), ... | | Authors: | Hino, T, Omura, N.S, Nakagawa, R, Togashi, T, Takeda, N.S, Hiramoto, T, Tasaka, S, Hirano, H, Tokuyama, T, Uosaki, H, Ishiguro, H, Yamano, H, Ozaki, Y, Motooka, D, Mori, H, Kirita, Y, Kise, Y, Itoh, Y, Matoba, S, Aburatani, H, Yachie, N, Siksnys, V, Ohmori, T, Hoshino, A, Nureki, O. | | Deposit date: | 2023-04-13 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Minimal and most efficient genome editing Cas enzyme
To Be Published
|
|
351D
 
 | |
168D
 
 | |
3MVD
 
 | | Crystal structure of the chromatin factor RCC1 in complex with the nucleosome core particle | | Descriptor: | DNA (146-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Makde, R.D, England, J.R, Yennawar, H.P, Tan, S. | | Deposit date: | 2010-05-04 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of RCC1 chromatin factor bound to the nucleosome core particle.
Nature, 467, 2010
|
|
6QLD
 
 | | Structure of inner kinetochore CCAN-Cenp-A complex | | Descriptor: | DNA (125-MER), Histone H2A.1, Histone H2B.1, ... | | Authors: | Yan, K, Yang, J, Zhang, Z, McLaughlin, S.H, Chang, L, Fasci, D, Heck, A.J.R, Barford, D. | | Deposit date: | 2019-01-31 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Structure of the inner kinetochore CCAN complex assembled onto a centromeric nucleosome.
Nature, 574, 2019
|
|
