6Q2O
 
 | | Cryo-EM structure of RET/GFRa2/NRTN extracellular complex. The 3D refinement was applied with C2 symmetry. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GDNF family receptor alpha-2, ... | | Authors: | Li, J, Shang, G.J, Chen, Y.J, Brautigam, C.A, Liou, J, Zhang, X.W, Bai, X.C. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Cryo-EM analyses reveal the common mechanism and diversification in the activation of RET by different ligands.
Elife, 8, 2019
|
|
6Q2N
 
 | | Cryo-EM structure of RET/GFRa1/GDNF extracellular complex | | Descriptor: | CALCIUM ION, GDNF family receptor alpha-1, Glial cell line-derived neurotrophic factor, ... | | Authors: | Li, J, Shang, G.J, Chen, Y.J, Brautigam, C.A, Liou, J, Zhang, X.W, Bai, X.C. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM analyses reveal the common mechanism and diversification in the activation of RET by different ligands.
Elife, 8, 2019
|
|
5JLG
 
 | | The X-ray structure of the adduct formed in the reaction between bovine pancreatic ribonuclease and compound I, a piano-stool organometallic Ru(II) arene compound containing an O,S-chelating ligand | | Descriptor: | DIMETHYL SULFOXIDE, RUTHENIUM ION, Ribonuclease pancreatic, ... | | Authors: | Ferraro, G, Merlino, A. | | Deposit date: | 2016-04-27 | | Release date: | 2016-08-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Unusual mode of protein binding by a cytotoxic pi-arene ruthenium(ii) piano-stool compound containing an O,S-chelating ligand.
Dalton Trans, 45, 2016
|
|
8VUW
 
 | | ELIC5 with cysteamine in 2:1:1 POPC:POPE:POPG nanodisc in open conformation | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, 2-AMINO-ETHANETHIOL, Erwinia chrysanthemi ligand-gated ion channel | | Authors: | Petroff II, J.T, Deng, Z, Rau, M.J, Fitzpatrick, J.A.J, Yuan, P, Cheng, W.W.L. | | Deposit date: | 2024-01-29 | | Release date: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Open-channel structure of a pentameric ligand-gated ion channel reveals a mechanism of leaflet-specific phospholipid modulation.
Nat Commun, 13, 2022
|
|
8D64
 
 | | ELIC with cysteamine in POPC nanodisc | | Descriptor: | 2-AMINO-ETHANETHIOL, Erwinia ligand-gated ion channel | | Authors: | Petroff II, J.T, Deng, Z, Rau, M.J, Fitzpatrick, J.A.J, Yuan, P, Cheng, W.W.L. | | Deposit date: | 2022-06-06 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Open-channel structure of a pentameric ligand-gated ion channel reveals a mechanism of leaflet-specific phospholipid modulation.
Nat Commun, 13, 2022
|
|
8D67
 
 | | ELIC3 with cysteamine in 2:1:1 POPC:POPE:POPG nanodisc | | Descriptor: | 2-AMINO-ETHANETHIOL, Erwinia ligand-gated ion channel | | Authors: | Petroff II, J.T, Deng, Z, Rau, M.J, Fitzpatrick, J.A.J, Yuan, P, Cheng, W.W.L. | | Deposit date: | 2022-06-06 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Open-channel structure of a pentameric ligand-gated ion channel reveals a mechanism of leaflet-specific phospholipid modulation.
Nat Commun, 13, 2022
|
|
8D65
 
 | | ELIC apo in 2:1:1 POPC:POPE:POPG nanodisc | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Erwinia ligand-gated ion channel | | Authors: | Petroff II, J.T, Deng, Z, Rau, M.J, Fitzpatrick, J.A.J, Yuan, P, Cheng, W.W.L. | | Deposit date: | 2022-06-06 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Open-channel structure of a pentameric ligand-gated ion channel reveals a mechanism of leaflet-specific phospholipid modulation.
Nat Commun, 13, 2022
|
|
8D63
 
 | | ELIC apo in POPC nanodisc | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Erwinia ligand-gated ion channel | | Authors: | Petroff II, J.T, Deng, Z, Rau, M.J, Fitzpatrick, J.A.J, Yuan, P, Cheng, W.W.L. | | Deposit date: | 2022-06-06 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Open-channel structure of a pentameric ligand-gated ion channel reveals a mechanism of leaflet-specific phospholipid modulation.
Nat Commun, 13, 2022
|
|
8D66
 
 | | ELIC with cysteamine in 2:1:1 POPC:POPE:POPG nanodisc | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, 2-AMINO-ETHANETHIOL, Erwinia ligand-gated ion channel | | Authors: | Petroff II, J.T, Deng, Z, Rau, M.J, Fitzpatrick, J.A.J, Yuan, P, Cheng, W.W.L. | | Deposit date: | 2022-06-06 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Open-channel structure of a pentameric ligand-gated ion channel reveals a mechanism of leaflet-specific phospholipid modulation.
Nat Commun, 13, 2022
|
|
6Q2J
 
 | | Cryo-EM structure of extracellular dimeric complex of RET/GFRAL/GDF15 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GDNF family receptor alpha-like, ... | | Authors: | Li, J, Shang, G.J, Chen, Y.J, Brautigam, C.A, Liou, J, Zhang, X.W, Bai, X.C. | | Deposit date: | 2019-08-08 | | Release date: | 2019-10-02 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM analyses reveal the common mechanism and diversification in the activation of RET by different ligands.
Elife, 8, 2019
|
|
8TWV
 
 | | ELIC5 with Propylamine in spNW15 nanodiscs with 2:1:1 POPC:POPE:POPG | | Descriptor: | 3-AMINOPROPANE, Gamma-aminobutyric-acid receptor subunit beta-1 | | Authors: | Dalal, V, Arcario, M.J, Petroff II, J.T, Deitzen, N.M, Tan, B.K, Brannigan, G, Cheng, W.W.L. | | Deposit date: | 2023-08-21 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Lipid nanodisc scaffold and size alter the structure of a pentameric ligand-gated ion channel.
Nat Commun, 15, 2024
|
|
1LIH
 
 | | THREE-DIMENSIONAL STRUCTURES OF THE LIGAND-BINDING DOMAIN OF THE BACTERIAL ASPARTATE RECEPTOR WITH AND WITHOUT A LIGAND | | Descriptor: | 1,10-PHENANTHROLINE, ASPARTATE RECEPTOR | | Authors: | Kim, S.-H, Scott, W, Yeh, J.I, Prive, G.G, Milburn, M. | | Deposit date: | 1995-04-18 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand.
Science, 254, 1991
|
|
8T3V
 
 | | Cryo-EM structure of the DHA bound FFA1-Gq complex | | Descriptor: | CHOLESTEROL, DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, Free fatty acid receptor 1, ... | | Authors: | Zhang, X, Tikhonova, I, Milligan, G, Zhang, C. | | Deposit date: | 2023-06-07 | | Release date: | 2024-01-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural basis for the ligand recognition and signaling of free fatty acid receptors.
Sci Adv, 10, 2024
|
|
7UW2
 
 | | Crystal structure of human Retinoid X receptor alpha ligand binding domain complex with UAB116 and coactivator peptide GRIP-1 | | Descriptor: | (2E,4E,6Z,8E)-8-{3-[(2S)-butan-2-yl]-2-(3-methylbutyl)cyclohex-2-en-1-ylidene}-3,7-dimethylocta-2,4,6-trienoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Chattopadhyay, D, Yang, Z, Atigadda, V. | | Deposit date: | 2022-05-02 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Conformationally Defined Rexinoids for the Prevention of Inflammation and Nonmelanoma Skin Cancers.
J.Med.Chem., 65, 2022
|
|
7UW4
 
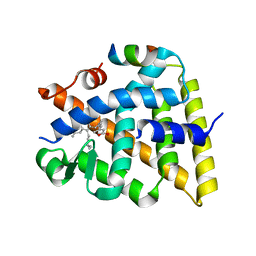 | | Crystal structure of human Retinoid X receptor alpha ligand binding domain complex with UAB113 and coactivator peptide GRIP-1 | | Descriptor: | (2E,4E,6Z,8E)-3,7-dimethyl-8-[2-(3-methylbutyl)-3-propylcyclohex-2-en-1-ylidene]octa-2,4,6-trienoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Chattopadhyay, D, Yang, Z, Atigadda, V. | | Deposit date: | 2022-05-02 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformationally Defined Rexinoids for the Prevention of Inflammation and Nonmelanoma Skin Cancers.
J.Med.Chem., 65, 2022
|
|
1Z17
 
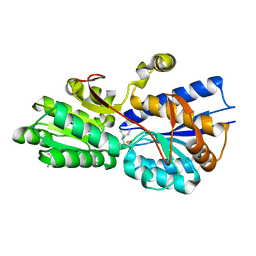 | | Crystal structure analysis of periplasmic Leu/Ile/Val-binding protein with bound ligand isoleucine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ISOLEUCINE, Leu/Ile/Val-binding protein | | Authors: | Trakhanov, S.D, Vyas, N.K, Kristensen, D.M, Ma, J, Quiocho, F.A. | | Deposit date: | 2005-03-03 | | Release date: | 2005-10-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Ligand-free and -bound structures of the binding protein (LivJ) of the Escherichia coli ABC leucine/isoleucine/valine transport system: trajectory and dynamics of the interdomain rotation and ligand specificity.
Biochemistry, 44, 2005
|
|
4EB6
 
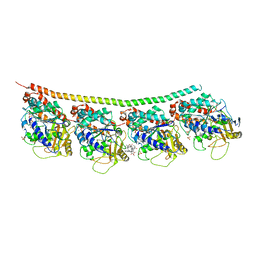 | | Tubulin-Vinblastine: Stathmin-like complex | | Descriptor: | (2ALPHA,2'BETA,3BETA,4ALPHA,5BETA)-VINCALEUKOBLASTINE, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Ranaivoson, F.M, Gigant, B, Knossow, M. | | Deposit date: | 2012-03-23 | | Release date: | 2012-08-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | Structural plasticity of tubulin assembly probed by vinca-domain ligands.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1AJQ
 
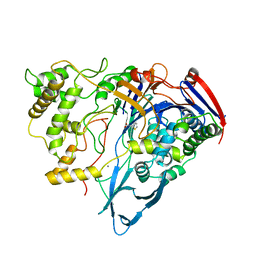 | | PENICILLIN ACYLASE COMPLEXED WITH THIOPHENEACETIC ACID | | Descriptor: | CALCIUM ION, PENICILLIN AMIDOHYDROLASE, THIOPHENEACETIC ACID | | Authors: | Done, S.H. | | Deposit date: | 1997-05-07 | | Release date: | 1997-11-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Ligand-induced conformational change in penicillin acylase.
J.Mol.Biol., 284, 1998
|
|
1AJP
 
 | |
3DU7
 
 | | Tubulin-colchicine-phomopsin A: Stathmin-like domain complex | | Descriptor: | 2-MERCAPTO-N-[1,2,3,10-TETRAMETHOXY-9-OXO-5,6,7,9-TETRAHYDRO-BENZO[A]HEPTALEN-7-YL]ACETAMIDE, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Cormier, A, Marchand, M, Ravelli, R.B, Knossow, M, Gigant, B. | | Deposit date: | 2008-07-17 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structural insight into the inhibition of tubulin by vinca domain peptide ligands
Embo Rep., 9, 2008
|
|
8T3O
 
 | | Cryo-EM structure of the TUG-891 bound FFA4-Gq complex | | Descriptor: | (2R)-1-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, 3-{4-[(4-fluoro-4'-methyl[1,1'-biphenyl]-2-yl)methoxy]phenyl}propanoic acid, Free fatty acid receptor 4, ... | | Authors: | Zhang, X, Tikhonova, I, Milligan, G, Zhang, C. | | Deposit date: | 2023-06-07 | | Release date: | 2024-01-17 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis for the ligand recognition and signaling of free fatty acid receptors.
Sci Adv, 10, 2024
|
|
8T3Q
 
 | | Cryo-EM structure of the DHA bound FFA4-Gq complex | | Descriptor: | DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, X, Tikhonova, I, Milligan, G, Zhang, C. | | Deposit date: | 2023-06-07 | | Release date: | 2024-01-24 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural basis for the ligand recognition and signaling of free fatty acid receptors.
Sci Adv, 10, 2024
|
|
8T3S
 
 | | Cryo-EM structure of the Butyrate bound FFA2-Gq complex | | Descriptor: | CHOLESTEROL, Free fatty acid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, X, Tikhonova, I, Milligan, G, Zhang, C. | | Deposit date: | 2023-06-07 | | Release date: | 2024-01-24 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis for the ligand recognition and signaling of free fatty acid receptors.
Sci Adv, 10, 2024
|
|
1AI5
 
 | | PENICILLIN ACYLASE COMPLEXED WITH M-NITROPHENYLACETIC ACID | | Descriptor: | 2-(3-NITROPHENYL)ACETIC ACID, CALCIUM ION, PENICILLIN AMIDOHYDROLASE | | Authors: | Done, S.H. | | Deposit date: | 1997-05-01 | | Release date: | 1997-11-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Ligand-induced conformational change in penicillin acylase.
J.Mol.Biol., 284, 1998
|
|
1AI4
 
 | |
