301D
 
 | | CAPTURING THE STRUCTURE OF A CATALYTIC RNA INTERMEDIATE: RNA HAMMERHEAD RIBOZYME, MG(II)-SOAKED | | Descriptor: | MAGNESIUM ION, RNA HAMMERHEAD RIBOZYME | | Authors: | Scott, W.G, Murray, J.B, Arnold, J.R.P, Stoddard, B.L, Klug, A. | | Deposit date: | 1996-12-14 | | Release date: | 1997-01-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Capturing the structure of a catalytic RNA intermediate: the hammerhead ribozyme.
Science, 274, 1996
|
|
6UJE
 
 | | Crystal structure of the Clostridial cellulose synthase subunit Z (CcsZ) from Clostridioides difficile | | Descriptor: | CALCIUM ION, Endoglucanase | | Authors: | Scott, W, Lowrance, B, Anderson, A.C, Weadge, J.T. | | Deposit date: | 2019-10-03 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification of the Clostridial cellulose synthase and characterization of the cognate glycosyl hydrolase, CcsZ.
Plos One, 15, 2020
|
|
6UJF
 
 | | Crystal structure of the Clostridial cellulose synthase subunit Z (CcsZ) from Clostridioides difficile | | Descriptor: | Endoglucanase, beta-D-glucopyranose | | Authors: | Scott, W, Lowrance, B, Anderson, A.C, Weadge, J.T. | | Deposit date: | 2019-10-03 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of the Clostridial cellulose synthase and characterization of the cognate glycosyl hydrolase, CcsZ.
Plos One, 15, 2020
|
|
299D
 
 | | CAPTURING THE STRUCTURE OF A CATALYTIC RNA INTERMEDIATE: THE HAMMERHEAD RIBOZYME | | Descriptor: | RNA HAMMERHEAD RIBOZYME | | Authors: | Scott, W.G, Murray, J.B, Arnold, J.R.P, Stoddard, B.L, Klug, A. | | Deposit date: | 1996-12-14 | | Release date: | 1997-01-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Capturing the structure of a catalytic RNA intermediate: the hammerhead ribozyme.
Science, 274, 1996
|
|
300D
 
 | | CAPTURING THE STRUCTURE OF A CATALYTIC RNA INTERMEDIATE: RNA HAMMERHEAD RIBOZYME, MN(II)-SOAKED | | Descriptor: | MANGANESE (II) ION, RNA HAMMERHEAD RIBOZYME | | Authors: | Scott, W.G, Murray, J.B, Arnold, J.R.P, Stoddard, B.L, Klug, A. | | Deposit date: | 1996-12-14 | | Release date: | 1997-01-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Capturing the structure of a catalytic RNA intermediate: the hammerhead ribozyme.
Science, 274, 1996
|
|
1MME
 
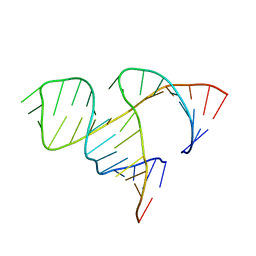 | |
3ZD4
 
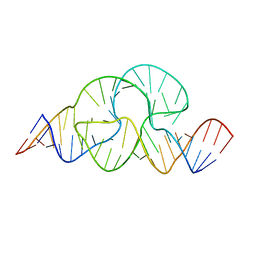 | |
2XKM
 
 | | Consensus structure of Pf1 filamentous bacteriophage from X-ray fibre diffraction and solid-state NMR | | Descriptor: | CAPSID PROTEIN G8P | | Authors: | Straus, S.K, P Scott, W.R, Schwieters, C.D, Marvin, D.A. | | Deposit date: | 2010-07-09 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-20 | | Method: | FIBER DIFFRACTION (3.3 Å), SOLID-STATE NMR | | Cite: | Consensus Structure of Pf1 Filamentous Bacteriophage from X-Ray Fibre Diffraction and Solid-State NMR.
Eur.Biophys.J., 40, 2011
|
|
359D
 
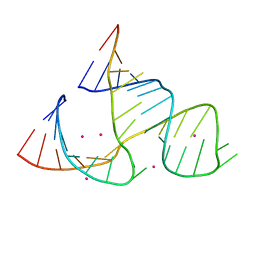 | |
2LIG
 
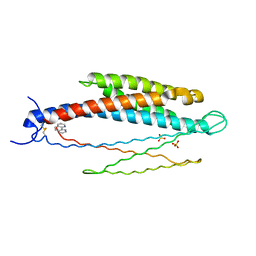 | | THREE-DIMENSIONAL STRUCTURES OF THE LIGAND-BINDING DOMAIN OF THE BACTERIAL ASPARTATE RECEPTOR WITH AND WITHOUT A LIGAND | | Descriptor: | 1,10-PHENANTHROLINE, ASPARTATE RECEPTOR, ASPARTIC ACID, ... | | Authors: | Kim, S.-H, Yeh, J.I, Prive, G.G, Milburn, M, Scott, W, Koshland Junior, D.E. | | Deposit date: | 1995-04-18 | | Release date: | 1995-09-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand.
Science, 254, 1991
|
|
5G2N
 
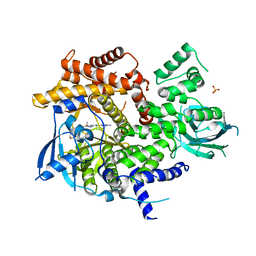 | | X-ray structure of PI3Kinase Gamma in complex with Copanlisib | | Descriptor: | 2-azanyl-~{N}-[7-methoxy-8-(3-morpholin-4-ylpropoxy)-2,3-dihydroimidazo[1,2-c]quinazolin-5-yl]pyrimidine-5-carboxamide, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM, SULFATE ION | | Authors: | Schaefer, M, Scott, W.J, Hentemann, M.F, Rowley, R.B, Bull, C.O, Jenkins, S, Bullion, A.M, Johnson, J, Redman, A, Robbins, A.H, Esler, W, Fracasso, R.P, Garrison, T, Hamilton, M, Michels, M, Wood, J.E, Wilkie, D.P, Xiao, H, Levy, J, Liu, N, Stasik, E, Brands, M, Lefranc, J. | | Deposit date: | 2016-04-11 | | Release date: | 2016-04-20 | | Last modified: | 2016-08-10 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Discovery and Sar of Novel 2,3-Dihydroimidazo(1,2-C)Quinazoline Pi3K Inhibitors: Identification of Copanlisib (Bay 80-6946)
Chemmedchem, 11, 2016
|
|
4RNE
 
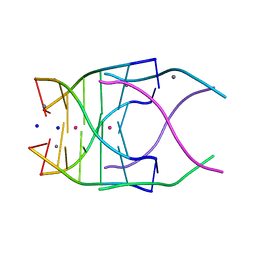 | | Structural variations and solvent structure of UGGGGU quadruplexes stabilized by Sr2+ ions | | Descriptor: | CALCIUM ION, RNA (5'-R(*UP*GP*GP*GP*GP*U)-3'), SODIUM ION, ... | | Authors: | Fyfe, A.C, Dunten, P.W, Scott, W.G. | | Deposit date: | 2014-10-24 | | Release date: | 2014-11-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Structural Variations and Solvent Structure of r(UGGGGU) Quadruplexes Stabilized by Sr(2+) Ions.
J.Mol.Biol., 427, 2015
|
|
1LIH
 
 | | THREE-DIMENSIONAL STRUCTURES OF THE LIGAND-BINDING DOMAIN OF THE BACTERIAL ASPARTATE RECEPTOR WITH AND WITHOUT A LIGAND | | Descriptor: | 1,10-PHENANTHROLINE, ASPARTATE RECEPTOR | | Authors: | Kim, S.-H, Scott, W, Yeh, J.I, Prive, G.G, Milburn, M. | | Deposit date: | 1995-04-18 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand.
Science, 254, 1991
|
|
2QUW
 
 | |
2VMK
 
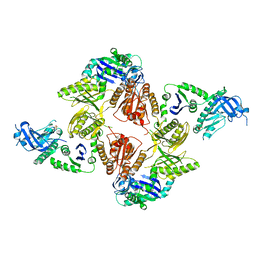 | | Crystal Structure of E. coli RNase E Apoprotein - Catalytic Domain | | Descriptor: | RIBONUCLEASE E, SULFATE ION, ZINC ION | | Authors: | Koslover, D.J, Callaghan, A.J, Marcaida, M.J, Martick, M, Scott, W.G, Luisi, B.F. | | Deposit date: | 2008-01-28 | | Release date: | 2008-07-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The Crystal Structure of the Escherichia Coli Rnase E Apoprotein and a Mechanism for RNA Degradation.
Structure, 16, 2008
|
|
2VRT
 
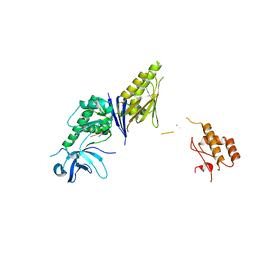 | | Crystal Structure of E. coli RNase E possessing M1 RNA fragments - Catalytic Domain | | Descriptor: | 5'-R(*UP*UP)-3', 5'-R(*UP*UP*GP)-3', RIBONUCLEASE E, ... | | Authors: | Koslover, D.J, Callaghan, A.J, Marcaida, M.J, Garman, E.F, Martick, M, Scott, W.G, Luisi, B.F. | | Deposit date: | 2008-04-14 | | Release date: | 2008-07-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The Crystal Structure of the Escherichia Coli Rnase E Apoprotein and a Mechanism for RNA Degradation.
Structure, 16, 2008
|
|
4RJ1
 
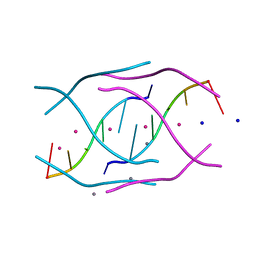 | | Structural variations and solvent structure of UGGGGU quadruplexes stabilized by Sr2+ ions | | Descriptor: | CALCIUM ION, RNA (5'-R(*UP*GP*GP*GP*GP*U)-3'), SODIUM ION, ... | | Authors: | Fyfe, A.C, Dunten, P.W, Scott, W.G. | | Deposit date: | 2014-10-08 | | Release date: | 2014-11-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Structural Variations and Solvent Structure of r(UGGGGU) Quadruplexes Stabilized by Sr(2+) Ions.
J.Mol.Biol., 427, 2015
|
|
4RKV
 
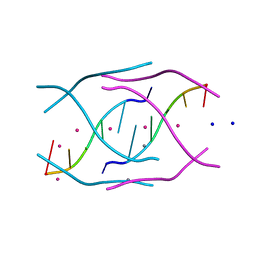 | | Structural variations and solvent structure of UGGGGU quadruplexes stabilized by Sr2+ ions | | Descriptor: | CALCIUM ION, RNA (5'-R(*UP*GP*GP*GP*GP*U)-3'), SODIUM ION, ... | | Authors: | Fyfe, A.C, Dunten, P.W, Scott, W.G. | | Deposit date: | 2014-10-14 | | Release date: | 2014-11-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (0.88 Å) | | Cite: | Structural Variations and Solvent Structure of r(UGGGGU) Quadruplexes Stabilized by Sr(2+) Ions.
J.Mol.Biol., 427, 2015
|
|
5UNE
 
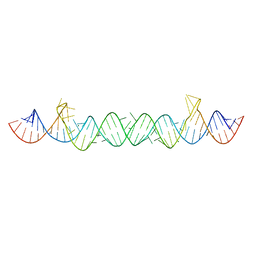 | |
1XJR
 
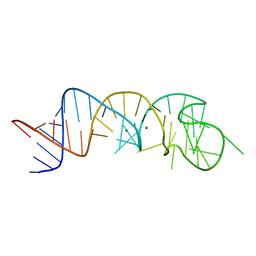 | | The Structure of a Rigorously Conserved RNA Element Within the SARS Virus Genome | | Descriptor: | MAGNESIUM ION, s2m RNA | | Authors: | Robertson, M.P, Igel, H, Baertsch, R, Haussler, D, Ares Jr, M, Scott, W.G. | | Deposit date: | 2004-09-24 | | Release date: | 2005-02-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of a rigorously conserved RNA element within the SARS virus genome
Plos Biol., 3, 2005
|
|
1Y28
 
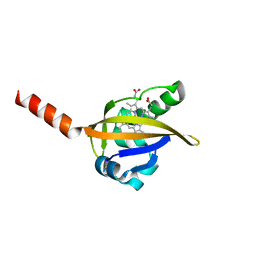 | | Crystal structure of the R220A metBJFIXL HEME domain | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Sensor protein fixL | | Authors: | Dunham, C.M, Dioum, E.M, Tuckerman, J.R, Gonzalez, G, Scott, W.G, Gilles-Gonzalez, M.A. | | Deposit date: | 2004-11-21 | | Release date: | 2004-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A distal arginine in the oxygen-sensing heme-PAS domains is essential to ligand binding, signal transduction, and structure
Biochemistry, 42, 2003
|
|
2BX2
 
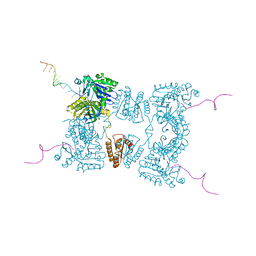 | | Catalytic domain of E. coli RNase E | | Descriptor: | MAGNESIUM ION, RIBONUCLEASE E, RNA (5'-R(*UP*UP*UP*AP*CP*AP*GP*UP*AP*UP*UP* UP*GP*UP*U)-3'), ... | | Authors: | Marcaida, M.J, Callaghan, A.J, Scott, W.G, Luisi, B.F. | | Deposit date: | 2005-07-21 | | Release date: | 2005-10-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of E. Coli Rnase E Catalytic Domain and Implications for RNA Processing and Turnover
Nature, 437, 2005
|
|
2C0X
 
 | | MOLECULAR STRUCTURE OF FD FILAMENTOUS BACTERIOPHAGE REFINED WITH RESPECT TO X-RAY FIBRE DIFFRACTION AND SOLID-STATE NMR DATA | | Descriptor: | COAT PROTEIN B | | Authors: | Marvin, D.A, Welsh, L.C, Symmons, M.F, Scott, W.R.P, Straus, S.K. | | Deposit date: | 2005-09-08 | | Release date: | 2005-12-14 | | Last modified: | 2020-07-29 | | Method: | SOLID-STATE NMR | | Cite: | Molecular Structure of Fd (F1, M13) Filamentous Bacteriophage Refined with Respect to X-Ray Fibre Diffraction and Solid-State NMR Data Supports Specific Models of Phage Assembly at the Bacterial Membrane.
J.Mol.Biol., 355, 2006
|
|
2C0B
 
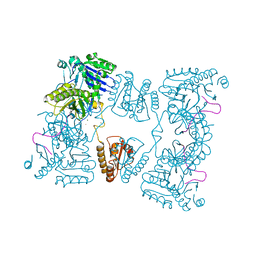 | | Catalytic domain of E. coli RNase E in complex with 13-mer RNA | | Descriptor: | 5'-R(*UP*UP*UP*AP*CP*AP*GP*UP*AP*UP*UP*UP*G)-3', MAGNESIUM ION, RIBONUCLEASE E, ... | | Authors: | Marcaida, M.J, Callaghan, A.J, Scott, W.G, Luisi, B.F. | | Deposit date: | 2005-08-30 | | Release date: | 2005-10-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Structure of E. Coli Rnase E Catalytic Domain and Implications for RNA Processing and Turnover
Nature, 437, 2005
|
|
2C0W
 
 | | Molecular Structure of fd Filamentous Bacteriophage Refined with Respect to X-ray Fibre Diffraction | | Descriptor: | COAT PROTEIN B | | Authors: | Marvin, D.A, Welsh, L.C, Symmons, M.F, Scott, W.R.P, Straus, S.K. | | Deposit date: | 2005-09-08 | | Release date: | 2005-12-14 | | Last modified: | 2024-02-14 | | Method: | FIBER DIFFRACTION (3.2 Å) | | Cite: | Molecular Structure of Fd (F1, M13) Filamentous Bacteriophage Refined with Respect to X-Ray Fibre Diffraction and Solid-State NMR Data Supports Specific Models of Phage Assembly at the Bacterial Membrane.
J.Mol.Biol., 355, 2006
|
|
