5ZOF
 
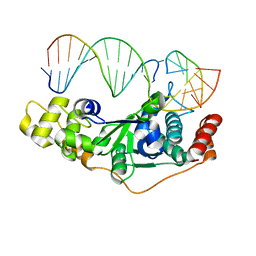 | | Crystal Structure of D181A/R192F hFen1 in complex with DNA | | Descriptor: | DNA (5'-D(*AP*CP*TP*TP*TP*GP*AP*GP*GP*CP*AP*GP*AP*G)-3'), DNA (5'-D(*CP*CP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*G)-3'), DNA (5'-D(*GP*CP*CP*CP*GP*TP*CP*C)-3'), ... | | Authors: | Han, W, Hua, Y, Zhao, Y. | | Deposit date: | 2018-04-13 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | Structural basis of 5' flap recognition and protein-protein interactions of human flap endonuclease 1.
Nucleic Acids Res., 46, 2018
|
|
5Z2V
 
 | | Crystal structure of RecR from Pseudomonas aeruginosa PAO1 | | Descriptor: | ANY 5'-MONOPHOSPHATE NUCLEOTIDE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Che, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2018-01-04 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of RecR, a member of the RecFOR DNA-repair pathway, from Pseudomonas aeruginosa PAO1.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
1VT5
 
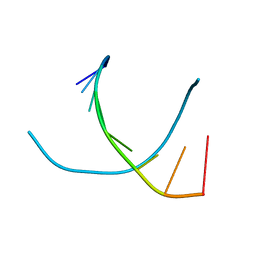 | |
8U0U
 
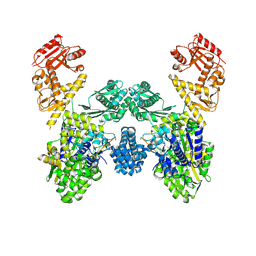 | | DdmD dimer in complex with ssDNA | | Descriptor: | DNA (5'-D(P*TP*TP*AP*TP*TP*TP*TP*TP*TP*TP*T)-3'), Helicase/UvrB N-terminal domain-containing protein | | Authors: | Bravo, J.P.K. | | Deposit date: | 2023-08-29 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Plasmid targeting and destruction by the DdmDE bacterial defence system.
Nature, 630, 2024
|
|
1V0D
 
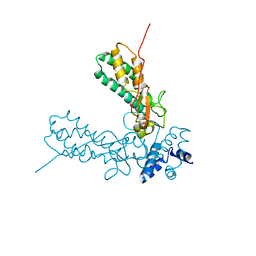 | | Crystal Structure of Caspase-activated DNase (CAD) | | Descriptor: | DNA FRAGMENTATION FACTOR 40 KDA SUBUNIT, LEAD (II) ION, MAGNESIUM ION, ... | | Authors: | Woo, E.-J, Kim, Y.-G, Kim, M.-S, Han, W.-D, Shin, S, Oh, B.-H. | | Deposit date: | 2004-03-26 | | Release date: | 2004-05-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Mechanism for Inactivation and Activation of Cad/Dff40 in the Apoptotic Pathway
Mol.Cell, 14, 2004
|
|
6M9M
 
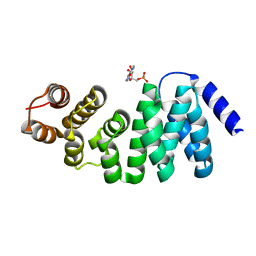 | |
6HP7
 
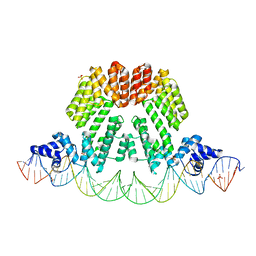 | |
8U0J
 
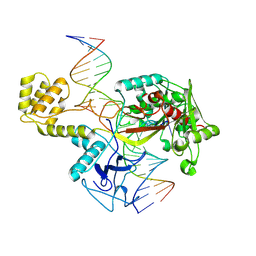 | | DdmE in complex with guide and target DNA | | Descriptor: | DNA (5'-D(P*AP*CP*TP*TP*AP*AP*AP*GP*TP*CP*TP*AP*AP*CP*CP*TP*AP*TP*AP*GP*GP*AP*T)-3'), DNA (5'-D(P*AP*TP*CP*CP*TP*AP*TP*AP*GP*GP*A)-3'), DNA (5'-D(P*GP*TP*TP*AP*GP*AP*CP*TP*TP*TP*AP*AP*GP*T)-3'), ... | | Authors: | Bravo, J.P.K. | | Deposit date: | 2023-08-29 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Plasmid targeting and destruction by the DdmDE bacterial defence system.
Nature, 630, 2024
|
|
8R0S
 
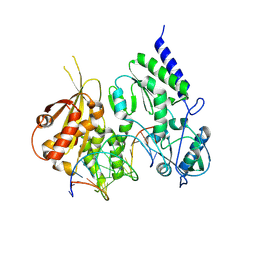 | | Structure of reverse transcriptase from Cauliflower Mosaic Virus in complex with RNA/DNA hybrid | | Descriptor: | DNA (5'-D(*GP*CP*TP*AP*CP*GP*CP*AP*CP*TP*GP*CP*TP*GP*GP*A)-3'), Enzymatic polyprotein, RNA (5'-R(*GP*UP*CP*CP*AP*GP*CP*AP*GP*UP*GP*CP*GP*UP*AP*GP*C)-3') | | Authors: | Prabaharan, C, Figiel, M, Chamera, S, Szczepanowski, R, Nowak, E, Nowotny, M. | | Deposit date: | 2023-10-31 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and biochemical characterization of cauliflower mosaic virus reverse transcriptase.
J.Biol.Chem., 300, 2024
|
|
8U0W
 
 | | DdmD monomer in complex with ssDNA | | Descriptor: | DNA (5'-D(P*TP*TP*AP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), Helicase/UvrB N-terminal domain-containing protein | | Authors: | Bravo, J.P.K. | | Deposit date: | 2023-08-29 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Plasmid targeting and destruction by the DdmDE bacterial defence system.
Nature, 630, 2024
|
|
2N9F
 
 | | Glucose as non natural nucleobase | | Descriptor: | DNA (5'-D(*CP*TP*AP*GP*CP*GP*GP*TP*CP*AP*TP*C)-3'), DNA (5'-D(*GP*AP*TP*GP*AP*CP*(4JA)P*GP*CP*TP*AP*G)-3') | | Authors: | Gomez-Pinto, I, Vengut-Climent, E, Lucas, R, Avino, A, Eritja, R, Gonzalez-Ibanez, C, Morales, J, Penalver, P, Fonseca-Guerra, C, Bickelhaupt, M. | | Deposit date: | 2015-11-20 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Glucose-Nucleobase Pseudo Base Pairs: Biomolecular Interactions within DNA.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
1JYH
 
 | |
7CSZ
 
 | | Crystal structure of the N-terminal tandem RRM domains of RBM45 in complex with single-stranded DNA | | Descriptor: | DNA (5'-D(*CP*GP*AP*CP*GP*GP*GP*AP*CP*GP*C)-3'), RNA-binding protein 45 | | Authors: | Chen, X, Yang, Z, Wang, W, Wang, M. | | Deposit date: | 2020-08-17 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for RNA recognition by the N-terminal tandem RRM domains of human RBM45.
Nucleic Acids Res., 49, 2021
|
|
7XI9
 
 | | Cryo-EM structure of human DNMT1 (aa:351-1616) in complex with ubiquitinated H3 and hemimethylated DNA analog (CXXC-ordered form) | | Descriptor: | DNA (5'-D(*AP*CP*TP*TP*AP*(5CM)P*GP*GP*AP*AP*GP*G)-3'), DNA (5'-D(*CP*CP*TP*TP*CP*(C55)P*GP*TP*AP*AP*GP*T)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Onoda, H, Kikuchi, A, Kori, S, Yoshimi, S, Yamagata, A, Arita, K. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-30 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Structural basis for activation of DNMT1.
Nat Commun, 13, 2022
|
|
7XIB
 
 | | Cryo-EM structure of human DNMT1 (aa:351-1616) in complex with ubiquitinated H3 and hemimethylated DNA analog (CXXC-disordered form) | | Descriptor: | DNA (5'-D(*AP*CP*TP*TP*AP*(5CM)P*GP*GP*AP*AP*GP*G)-3'), DNA (5'-D(*CP*CP*TP*TP*CP*(C55)P*GP*TP*AP*AP*GP*T)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Onoda, H, Kikuchi, A, Kori, S, Yoshimi, S, Yamagata, A, Arita, K. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-30 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.23 Å) | | Cite: | Structural basis for activation of DNMT1.
Nat Commun, 13, 2022
|
|
4JD8
 
 | | Racemic-[Ru(phen)2(dppz)]2+] bound to synthetic DNA at high resolution | | Descriptor: | DNA (5'-D(*AP*TP*GP*CP*AP*T)-3'), Delta-Ru(phen)2(dppz) complex, Lambda-Ru(phen)2(dppz) complex, ... | | Authors: | Cook, D.S, Hall, J.P, Cardin, C.J. | | Deposit date: | 2013-02-24 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | X-ray Crystal Structure of rac-[Ru(phen)2dppz](2+) with d(ATGCAT)2 Shows Enantiomer Orientations and Water Ordering.
J.Am.Chem.Soc., 135, 2013
|
|
1EEG
 
 | | A(GGGG)A HEXAD PAIRING ALIGMENT FOR THE D(G-G-A-G-G-A-G) SEQUENCE | | Descriptor: | DNA (5'-D(*GP*GP*AP*GP*GP*A)-3') | | Authors: | Kettani, A, Gorin, A, Majumdar, A, Hermann, T, Skripkin, E, Zhao, H, Jones, R, Patel, D.J. | | Deposit date: | 2000-01-31 | | Release date: | 2000-04-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A dimeric DNA interface stabilized by stacked A.(G.G.G.G).A hexads and coordinated monovalent cations.
J.Mol.Biol., 297, 2000
|
|
8DYK
 
 | |
2PIE
 
 | |
3MGV
 
 | | Cre recombinase-DNA transition state | | Descriptor: | DNA (5'-D(*CP*AP*TP*AP*TP*GP*CP*TP*AP*TP*AP*CP*GP*AP*AP*GP*TP*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*CP*TP*TP*CP*GP*TP*AP*TP*AP*G)-3'), Recombinase cre, ... | | Authors: | Gibb, B.P, Gupta, K, Ghosh, K, Sharp, R, Chen, J, Van Duyne, G.D. | | Deposit date: | 2010-04-07 | | Release date: | 2010-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Requirements for catalysis in the Cre recombinase active site.
Nucleic Acids Res., 38, 2010
|
|
4QBR
 
 | |
3PX7
 
 | |
401D
 
 | |
4I9V
 
 | | The atomic structure of 5-Hydroxymethyl 2'-deoxycitidine base paired with 2'-deoxyguanosine in Dickerson Drew Dodecamer | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(5HC)P*GP*CP*G)-3'), MAGNESIUM ION, SPERMINE (FULLY PROTONATED FORM) | | Authors: | Nocek, B, Szulik, M.W, Joachimiak, A, Stone, M.P. | | Deposit date: | 2012-12-05 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Differential stabilities and sequence-dependent base pair opening dynamics of watson-crick base pairs with 5-hydroxymethylcytosine, 5-formylcytosine, or 5-carboxylcytosine.
Biochemistry, 54, 2015
|
|
6M4J
 
 | | SspA in complex with cysteine | | Descriptor: | CYSTEINE, PYRIDOXAL-5'-PHOSPHATE, SspA complex protein | | Authors: | Liu, L, Gao, H. | | Deposit date: | 2020-03-07 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Analysis of an l-Cysteine Desulfurase from an Ssp DNA Phosphorothioation System.
Mbio, 11, 2020
|
|
