3F5G
 
 | | Crystal structure of death associated protein kinase in complex with ADP and Mg2+ | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Death-associated protein kinase 1, MAGNESIUM ION | | Authors: | McNamara, L.K, Watterson, D.M, Brunzelle, J.S. | | Deposit date: | 2008-11-03 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural insight into nucleotide recognition by human death-associated protein kinase.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3F5U
 
 | | Crystal structure of the death associated protein kinase in complex with AMPPNP and Mg2+ | | Descriptor: | Death-associated protein kinase 1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | McNamara, L.K, Watterson, D.M, Brunzelle, J.S. | | Deposit date: | 2008-11-04 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight into nucleotide recognition by human death-associated protein kinase.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3F9Q
 
 | |
6S0W
 
 | | The crystal structure of kanamycin B dioxygenase (KanJ) from Streptomyces kanamyceticus in complex with nickel and kanamycin B sulfate | | Descriptor: | (1R,2S,3S,4R,6S)-4,6-DIAMINO-3-[(3-AMINO-3-DEOXY-ALPHA-D-GLUCOPYRANOSYL)OXY]-2-HYDROXYCYCLOHEXYL 2,6-DIAMINO-2,6-DIDEOXY-ALPHA-D-GLUCOPYRANOSIDE, DI(HYDROXYETHYL)ETHER, Kanamycin B dioxygenase, ... | | Authors: | Mrugala, B, Niedzialkowska, E, Minor, W, Borowski, T. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | A study on the structure, mechanism, and biochemistry of kanamycin B dioxygenase (KanJ)-an enzyme with a broad range of substrates.
Febs J., 288, 2021
|
|
3CY6
 
 | | Crystal Structure of E18Q DJ-1 | | Descriptor: | Protein DJ-1 | | Authors: | Witt, A.C, Lakshminarasimhan, M, Remington, B.C, Hasim, S, Pozharski, E, Wilson, M.A. | | Deposit date: | 2008-04-25 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Cysteine pKa depression by a protonated glutamic acid in human DJ-1.
Biochemistry, 47, 2008
|
|
6ROA
 
 | | Crystal structure of V57G mutant of human cystatin C | | Descriptor: | Cystatin-C | | Authors: | Orlikowska, M, Behrendt, I, Borek, D, Otwinowski, Z, Skowron, P, Szymanska, A. | | Deposit date: | 2019-05-10 | | Release date: | 2019-08-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | NMR and crystallographic structural studies of the extremely stable monomeric variant of human cystatin C with single amino acid substitution.
Febs J., 287, 2020
|
|
6RA6
 
 | | Ferric murine neuroglobin Gly-loop44-47/F106A mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Exertier, C, Freda, I, Montemiglio, L.C, Savino, C, Vallone, B. | | Deposit date: | 2019-04-05 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Lack of orientation selectivity of the heme insertion in murine neuroglobin revealed by resonance Raman spectroscopy.
Febs J., 287, 2020
|
|
6RL5
 
 | |
6UE0
 
 | | Crystal structure of dihydrodipicolinate synthase from Klebsiella pneumoniae bound to pyruvate | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate synthase, CHLORIDE ION, SULFATE ION | | Authors: | Impey, R.E, Lee, M, Hawkins, D.A, Sutton, J.M, Panjikar, S, Perugini, M.A, Soares da Costa, T.P. | | Deposit date: | 2019-09-20 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Mis-annotations of a promising antibiotic target in high-priority gram-negative pathogens.
Febs Lett., 594, 2020
|
|
4ZSL
 
 | | MITOGEN ACTIVATED PROTEIN KINASE 7 IN COMPLEX WITH INHIBITOR | | Descriptor: | 3-amino-5-[(4-chlorophenyl)amino]-N-[(1S)-1-phenylethyl]-1H-1,2,4-triazole-1-carboxamide, GLYCEROL, Mitogen-activated protein kinase 7 | | Authors: | Ogg, D.J, Tucker, J. | | Deposit date: | 2015-05-13 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery of a novel allosteric inhibitor-binding site in ERK5: comparison with the canonical kinase hinge ATP-binding site.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
6LH1
 
 | | Crystal structure of a cysteine-pair mutant (Y113C-P190C) of a bacterial bile acid transporter trapped in an outward-facing conformation | | Descriptor: | 2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CITRIC ACID, Transporter, ... | | Authors: | Wang, X, Lyu, Y, Ji, Y, Sun, Z, Zhou, X. | | Deposit date: | 2019-12-06 | | Release date: | 2020-12-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.861 Å) | | Cite: | An engineered disulfide bridge traps and validates an outward-facing conformation in a bile acid transporter.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6ROR
 
 | | REP related 18-mer DNA | | Descriptor: | REP related 18-mer DNA from C. hominis, STRONTIUM ION | | Authors: | Kolenko, P, Svoboda, J, Schneider, B. | | Deposit date: | 2019-05-13 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural variability of CG-rich DNA 18-mers accommodating double T-T mismatches.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
5UYY
 
 | | Crystal structure of prephenate dehydrogenase tyrA from Bacillus anthracis in complex with L-tyrosine | | Descriptor: | Prephenate dehydrogenase, TYROSINE | | Authors: | Shabalin, I.G, Hou, J, Cymborowski, M.T, Kwon, K, Christendat, D, Gritsunov, A.O, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-24 | | Release date: | 2017-03-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and biochemical analysis of Bacillus anthracis prephenate dehydrogenase reveals an unusual mode of inhibition by tyrosine via the ACT domain.
Febs J., 287, 2020
|
|
3FIB
 
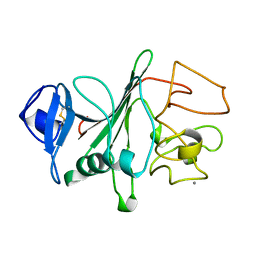 | | RECOMBINANT HUMAN GAMMA-FIBRINOGEN CARBOXYL TERMINAL FRAGMENT (RESIDUES 143-411) BOUND TO CALCIUM AT PH 6.0: A FURTHER REFINEMENT OF PDB ENTRY 1FIB, AND DIFFERS FROM 1FIB BY THE MODELLING OF A CIS PEPTIDE BOND BETWEEN RESIDUES K338 AND C339 | | Descriptor: | CALCIUM ION, FIBRINOGEN GAMMA CHAIN RESIDUES | | Authors: | Pratt, K.P, Cote, H.C.F, Chung, D.W, Stenkamp, R.E, Davie, E.W. | | Deposit date: | 1997-07-14 | | Release date: | 1997-09-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The primary fibrin polymerization pocket: three-dimensional structure of a 30-kDa C-terminal gamma chain fragment complexed with the peptide Gly-Pro-Arg-Pro.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
6S0U
 
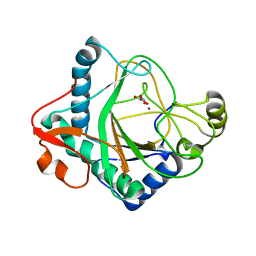 | | The crystal structure of kanamycin B dioxygenase (KanJ) from Streptomyces kanamyceticus in complex with nickel and 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mrugala, B, Porebski, P.J, Niedzialkowska, E, Minor, W, Borowski, T. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A study on the structure, mechanism, and biochemistry of kanamycin B dioxygenase (KanJ)-an enzyme with a broad range of substrates.
Febs J., 288, 2021
|
|
6ROS
 
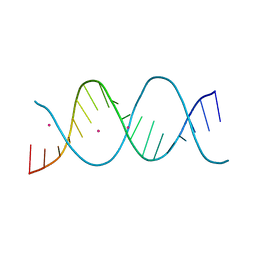 | | REP related 18-mer DNA | | Descriptor: | REP related 18-mer DNA from C. hominis, STRONTIUM ION | | Authors: | Kolenko, P, Svoboda, J, Schneider, B. | | Deposit date: | 2019-05-13 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural variability of CG-rich DNA 18-mers accommodating double T-T mismatches.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6LIS
 
 | | ASFV dUTPase in complex with dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, E165R | | Authors: | Liang, R, Peng, G.Q. | | Deposit date: | 2019-12-12 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural comparisons of host and African swine fever virus dUTPases reveal new clues for inhibitor development.
J.Biol.Chem., 296, 2020
|
|
6LJ3
 
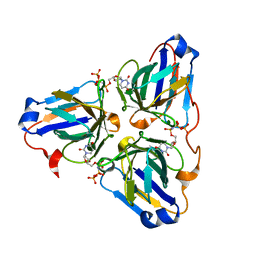 | |
6LJO
 
 | | African swine fever virus dUTPase | | Descriptor: | E165R | | Authors: | Liang, R, Peng, G.Q. | | Deposit date: | 2019-12-17 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural comparisons of host and African swine fever virus dUTPases reveal new clues for inhibitor development.
J.Biol.Chem., 296, 2020
|
|
6ROU
 
 | | REP related 18-mer DNA | | Descriptor: | REP related 18-mer DNA from H. parasuis, STRONTIUM ION | | Authors: | Kolenko, P, Svoboda, J, Schneider, B. | | Deposit date: | 2019-05-13 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Structural variability of CG-rich DNA 18-mers accommodating double T-T mismatches.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
5V0S
 
 | | Crystal structure of the ACT domain of prephenate dehydrogenase tyrA from Bacillus anthracis | | Descriptor: | CALCIUM ION, Prephenate dehydrogenase, SULFATE ION | | Authors: | Shabalin, I.G, Hou, J, Cymborowski, M.T, Otwinowski, Z, Kwon, K, Christendat, D, Gritsunov, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-28 | | Release date: | 2017-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and biochemical analysis of Bacillus anthracis prephenate dehydrogenase reveals an unusual mode of inhibition by tyrosine via the ACT domain.
Febs J., 287, 2020
|
|
2YZ3
 
 | | Crystallographic Investigation of Inhibition Mode of the VIM-2 Metallo-beta-lactamase from Pseudomonas aeruginosa with Mercaptocarboxylate Inhibitor | | Descriptor: | (S)-2-(MERCAPTOMETHYL)-5-PHENYLPENTANOIC ACID, Metallo-beta-lactamase, SULFATE ION, ... | | Authors: | Yamaguchi, Y, Yamagata, Y, Arakawa, Y, Kurosaki, H. | | Deposit date: | 2007-05-02 | | Release date: | 2008-03-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic investigation of the inhibition mode of a VIM-2 metallo-beta-lactamase from Pseudomonas aeruginosa by a mercaptocarboxylate inhibitor.
J.Med.Chem., 50, 2007
|
|
5VEH
 
 | | Re-refinement OF THE PDB STRUCTURE 1yiz of Aedes aegypti kynurenine aminotransferase | | Descriptor: | BROMIDE ION, GLYCEROL, Kynurenine aminotransferase | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-04 | | Release date: | 2017-11-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
7CA0
 
 | | Crystal structure of dihydroorotase in complex with 5-fluoroorotic acid from Saccharomyces cerevisiae | | Descriptor: | 5-FLUORO-2,6-DIOXO-1,2,3,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Dihydroorotase, ZINC ION | | Authors: | Guan, H.H, Huang, Y.H, Huang, C.Y, Chen, C.J. | | Deposit date: | 2020-06-08 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Complexed Crystal Structure of Saccharomyces cerevisiae Dihydroorotase with Inhibitor 5-Fluoroorotate Reveals a New Binding Mode.
Bioinorg Chem Appl, 2021, 2021
|
|
7CA1
 
 | | Crystal structure of dihydroorotase in complex with plumbagin from Saccharomyces cerevisiae | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 5-hydroxy-2-methylnaphthalene-1,4-dione, Dihydroorotase, ... | | Authors: | Guan, H.H, Huang, Y.H, Huang, C.Y, Chen, C.J. | | Deposit date: | 2020-06-08 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Plumbagin, a Natural Product with Potent Anticancer Activities, Binds to and Inhibits Dihydroorotase, a Key Enzyme in Pyrimidine Biosynthesis.
Int J Mol Sci, 22, 2021
|
|
