7Z4S
 
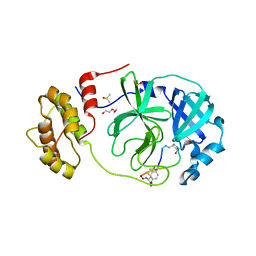 | | Crystal structure of SARS-CoV-2 Mpro in complex with cyclic peptide GM4 including unnatural amino acids. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Owen, C.D, Miura, T, Malla, T, Lukacik, L, Strain-Damerell, C.M, Tumber, A, Brewitz, L, McDonough, M.A, Salah, E, Terasaka, N, Katoh, T, Kawamura, A, Schofield, C.J, Suga, H, Walsh, M.A. | | Deposit date: | 2022-03-04 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In vitro selection of macrocyclic peptide inhibitors containing cyclic gamma 2,4 -amino acids targeting the SARS-CoV-2 main protease.
Nat.Chem., 15, 2023
|
|
7Z4N
 
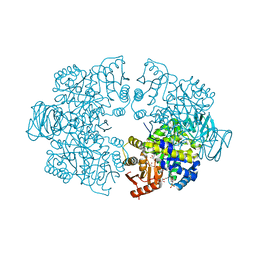 | | Plasmodium falciparum pyruvate kinase complexed with pyruvate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Dillenberger, M, Rahlfs, S, Becker, K, Fritz-Wolf, K. | | Deposit date: | 2022-03-04 | | Release date: | 2022-08-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Prominent role of cysteine residues C49 and C343 in regulating Plasmodium falciparum pyruvate kinase activity.
Structure, 30, 2022
|
|
7Z4M
 
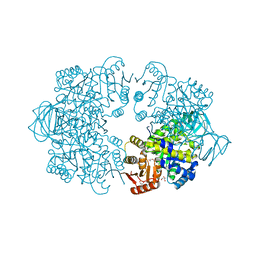 | | Plasmodium falciparum pyruvate kinase complexed with Mg2+ and K+ | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Dillenberger, M, Rahlfs, S, Becker, K, Fritz-Wolf, K. | | Deposit date: | 2022-03-04 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Prominent role of cysteine residues C49 and C343 in regulating Plasmodium falciparum pyruvate kinase activity.
Structure, 30, 2022
|
|
7Z3Y
 
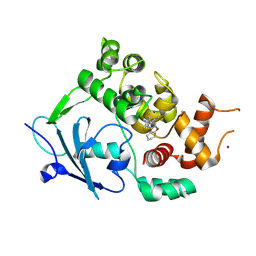 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH013545 | | Descriptor: | 2-[4-(3,5-dimethylpyrazol-1-yl)-2,6-bis(fluoranyl)phenyl]-~{N}-(4,5,6,7-tetrahydro-1,2-benzoxazol-3-yl)ethanamide, GLYCEROL, N-glycosylase/DNA lyase, ... | | Authors: | Scaletti, E.R, Stenmark, P. | | Deposit date: | 2022-03-02 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH013545
To Be Published
|
|
7Z3W
 
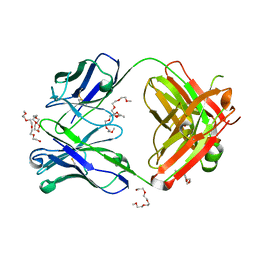 | | Crystal structure of the AAL160 Fab | | Descriptor: | 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, AAL160 Fab heavy-chain, AAL160 Fab light-chain | | Authors: | Rondeau, J.M, Lehmann, S. | | Deposit date: | 2022-03-02 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | "Redirecting an anti-IL-1 beta antibody to bind a new, unrelated and computationally predicted epitope on hIL-17A".
Commun Biol, 6, 2023
|
|
7Z3Q
 
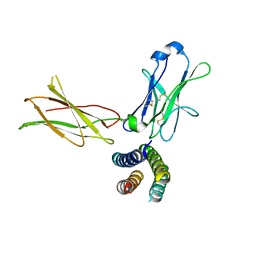 | | Crystal structure of the human leptin:LepR-CRH2 encounter complex to 3.6 A resolution. | | Descriptor: | Leptin, Leptin receptor | | Authors: | Verstraete, K, Verschueren, K, Alexandra, T, Savvides, S.N. | | Deposit date: | 2022-03-02 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.617 Å) | | Cite: | Mechanism of receptor assembly via the pleiotropic adipokine Leptin.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7Z3P
 
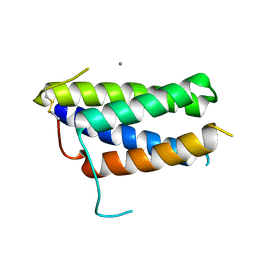 | | Crystal structure of the mouse leptin:LepR-CRH2 encounter complex to 1.95 A resolution. | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Leptin, ... | | Authors: | Tsirigotaki, A, Verschueren, K, Savvides, S.N, Verstraete, K. | | Deposit date: | 2022-03-02 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.943 Å) | | Cite: | Mechanism of receptor assembly via the pleiotropic adipokine Leptin.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7Z3L
 
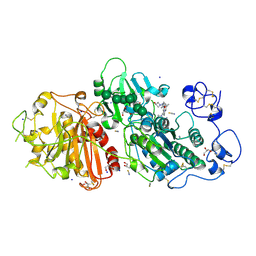 | | Autotaxin in complex with hybrid compound ziritaxestat (GLPG1690) | | Descriptor: | 2-[[2-ethyl-8-methyl-6-[4-[2-(3-oxidanylazetidin-1-yl)-2-oxidanylidene-ethyl]piperazin-1-yl]imidazo[1,2-a]pyridin-3-yl]-methyl-amino]-4-(4-fluorophenyl)-1,3-thiazole-5-carbonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Salgado-Polo, F, Ford, P, Heckmann, B, Perrakis, A. | | Deposit date: | 2022-03-02 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Autotaxin facilitates selective LPA receptor signaling.
Cell Chem Biol, 30, 2023
|
|
7Z3K
 
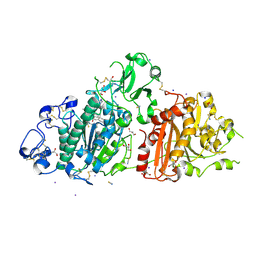 | | Autotaxin in complex with orthosteric site-binder CpdA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7alpha-hydroxycholesterol, CALCIUM ION, ... | | Authors: | Salgado-Polo, F, Ford, P, Heckmann, B, Perrakis, A. | | Deposit date: | 2022-03-02 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Autotaxin facilitates selective LPA receptor signaling.
Cell Chem Biol, 30, 2023
|
|
7Z3J
 
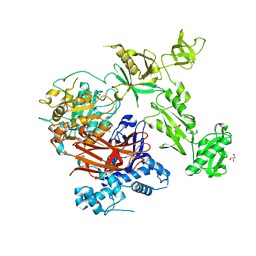 | | Structure of crystallisable rat Phospholipase C gamma 1 in complex with inositol 1,4,5-trisphosphate | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-1, CALCIUM ION, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, ... | | Authors: | Pinotsis, N, Bunney, T.D, Katan, M. | | Deposit date: | 2022-03-02 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of the membrane interactions of phospholipase C gamma reveals key features of the active enzyme.
Sci Adv, 8, 2022
|
|
7Z3I
 
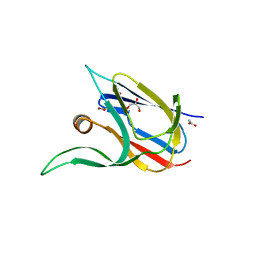 | | Crystal structure of the cupredoxin AcoP from Acidithiobacillus ferrooxidans, M171A mutant | | Descriptor: | ACETATE ION, AcoP, COPPER (II) ION, ... | | Authors: | Leone, P, Sciara, G, Ilbert, M. | | Deposit date: | 2022-03-02 | | Release date: | 2023-09-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Beyond the coupled distortion model: structural analysis of the single domain cupredoxin AcoP, a green mononuclear copper centre with original features.
Dalton Trans, 53, 2024
|
|
7Z3G
 
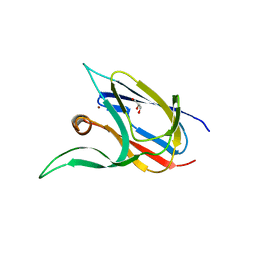 | | Crystal structure of the cupredoxin AcoP from Acidithiobacillus ferrooxidans, H166A mutant | | Descriptor: | AcoP, COPPER (I) ION, GLYCEROL | | Authors: | Leone, P, Sciara, G, Ilbert, M. | | Deposit date: | 2022-03-02 | | Release date: | 2023-09-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Beyond the coupled distortion model: structural analysis of the single domain cupredoxin AcoP, a green mononuclear copper centre with original features.
Dalton Trans, 53, 2024
|
|
7Z3F
 
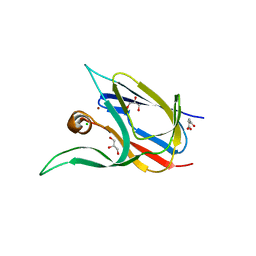 | | Crystal structure of the cupredoxin AcoP from Acidithiobacillus ferrooxidans, oxidized form | | Descriptor: | ACETATE ION, AcoP, CHLORIDE ION, ... | | Authors: | Leone, P, Sciara, G, Ilbert, M. | | Deposit date: | 2022-03-02 | | Release date: | 2023-09-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Beyond the coupled distortion model: structural analysis of the single domain cupredoxin AcoP, a green mononuclear copper centre with original features.
Dalton Trans, 53, 2024
|
|
7Z3B
 
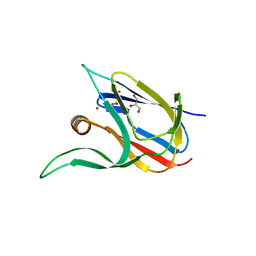 | | Crystal structure of the cupredoxin AcoP from Acidithiobacillus ferrooxidans, reduced form | | Descriptor: | ACETATE ION, AcoP, COPPER (I) ION, ... | | Authors: | Leone, P, Sciara, G, Ilbert, M. | | Deposit date: | 2022-03-02 | | Release date: | 2023-09-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Beyond the coupled distortion model: structural analysis of the single domain cupredoxin AcoP, a green mononuclear copper centre with original features.
Dalton Trans, 53, 2024
|
|
7Z39
 
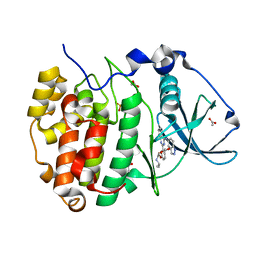 | | Structure of Belumosudil bound to CK2alpha | | Descriptor: | 2-[3-[4-(1~{H}-indazol-5-ylamino)quinazolin-2-yl]phenoxy]-~{N}-propan-2-yl-ethanamide, ACETATE ION, Casein kinase II subunit alpha, ... | | Authors: | Brear, P, Hyvonen, M. | | Deposit date: | 2022-03-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the Rho-associated coiled-coil kinase 2 inhibitor belumosudil bound to CK2 alpha.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
7Z38
 
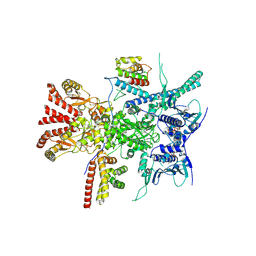 | | Structure of the RAF1-HSP90-CDC37 complex (RHC-I) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein HSP 90-beta, Hsp90 co-chaperone Cdc37, ... | | Authors: | Mesa, P, Garcia-Alonso, S, Barbacid, M, Montoya, G. | | Deposit date: | 2022-03-01 | | Release date: | 2022-09-14 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structure of the RAF1-HSP90-CDC37 complex reveals the basis of RAF1 regulation.
Mol.Cell, 82, 2022
|
|
7Z37
 
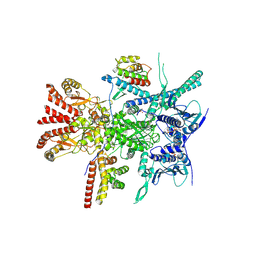 | | Structure of the RAF1-HSP90-CDC37 complex (RHC-II) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein HSP 90-beta, Hsp90 co-chaperone Cdc37, ... | | Authors: | Mesa, P, Garcia-Alonso, S, Barbacid, M, Montoya, G. | | Deposit date: | 2022-03-01 | | Release date: | 2022-09-14 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structure of the RAF1-HSP90-CDC37 complex reveals the basis of RAF1 regulation.
Mol.Cell, 82, 2022
|
|
7Z36
 
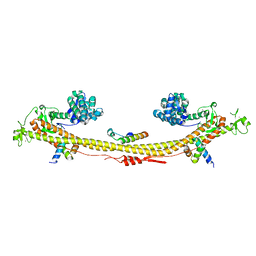 | | Crystal structure of the KAP1 tripartite motif in complex with the ZNF93 KRAB domain | | Descriptor: | Endolysin,Transcription intermediary factor 1-beta,Isoform 2 of Transcription intermediary factor 1-beta, SMARCAD1 CUE1 domain, ZINC ION, ... | | Authors: | Stoll, G.A, Modis, Y. | | Deposit date: | 2022-03-01 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and functional mapping of the KRAB-KAP1 repressor complex.
Embo J., 41, 2022
|
|
7Z35
 
 | |
7Z34
 
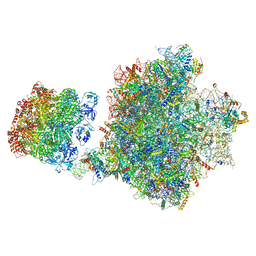 | | Structure of pre-60S particle bound to DRG1(AFG2). | | Descriptor: | 35S pre-ribosomal RNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Prattes, M, Grishkovskaya, I, Bergler, H, Haselbach, D. | | Deposit date: | 2022-03-01 | | Release date: | 2022-09-21 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Visualizing maturation factor extraction from the nascent ribosome by the AAA-ATPase Drg1.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7Z31
 
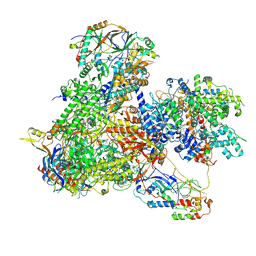 | | Structure of yeast RNA Polymerase III-Ty1 integrase complex at 2.7 A (focus subunit C11, no C11 C-terminal Zn-ribbon in the funnel pore). | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Nguyen, P.Q, Huecas, S, Plaza-Pegueroles, A, Fernandez-Tornero, C. | | Deposit date: | 2022-03-01 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural basis of Ty1 integrase tethering to RNA polymerase III for targeted retrotransposon integration.
Nat Commun, 14, 2023
|
|
7Z30
 
 | |
7Z2Z
 
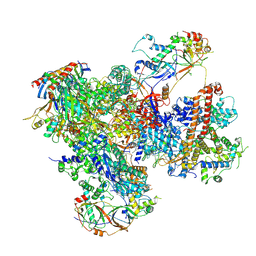 | | Structure of yeast RNA Polymerase III-DNA-Ty1 integrase complex (Pol III-DNA-IN1) at 3.1 A | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Nguyen, P.Q, Fernandez-Tornero, C. | | Deposit date: | 2022-03-01 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis of Ty1 integrase tethering to RNA polymerase III for targeted retrotransposon integration.
Nat Commun, 14, 2023
|
|
7Z2W
 
 | | Escherichia coli periplasmic phytase AppA D304A,T305E mutant, complex with myo-inositol hexakissulfate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Acidphosphatase, D-MYO-INOSITOL-HEXASULPHATE, ... | | Authors: | Acquistapace, I.M, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2022-03-01 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Insights to the Structural Basis for the Stereospecificity of the Escherichia coli Phytase, AppA.
Int J Mol Sci, 23, 2022
|
|
7Z2T
 
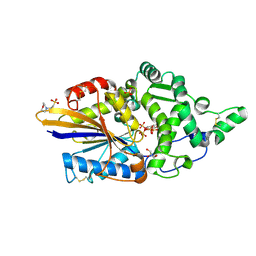 | | Escherichia coli periplasmic phytase AppA D304A mutant, complex with myo-inositol hexakissulfate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Acidphosphatase, D-MYO-INOSITOL-HEXASULPHATE, ... | | Authors: | Acquistapace, I.M, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2022-02-28 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Insights to the Structural Basis for the Stereospecificity of the Escherichia coli Phytase, AppA.
Int J Mol Sci, 23, 2022
|
|
