1UW0
 
 | |
2ZAS
 
 | | Crystal structure of human estrogen-related receptor gamma ligand binding domain complex with 4-alpha-cumylphenol, a bisphenol A derivative | | Descriptor: | 4-(1-methyl-1-phenylethyl)phenol, Estrogen-related receptor gamma, GLYCEROL | | Authors: | Matsushima, A, Kakuta, Y, Teramoto, T, Shimohigashi, Y. | | Deposit date: | 2007-10-09 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ERRgamma tethers strongly bisphenol A and 4-alpha-cumylphenol in an induced-fit manner
Biochem.Biophys.Res.Commun., 373, 2008
|
|
4V1Z
 
 | | The 3-D structure of the cellobiohydrolase, Cel7A, from Aspergillus fumigatus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLOBIOHYDROLASE, ZINC ION | | Authors: | Moroz, O.V, Maranta, M, Shaghasi, T, Harris, P.V, Wilson, K.S, Davies, G.J. | | Deposit date: | 2014-10-04 | | Release date: | 2015-01-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The Three-Dimensional Structure of the Cellobiohydrolase Cel7A from Aspergillus Fumigatus at 1.5 A Resolution
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4XM8
 
 | | Anthrax toxin lethal factor with ligand-induced binding pocket | | Descriptor: | Lethal factor, N-hydroxy-N~2~-{[3-(methoxymethyl)phenyl]sulfonyl}-N~2~-(2-methylpropyl)-D-valinamide, ZINC ION | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2015-01-14 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ligand-induced expansion of the S1' site in the anthrax toxin lethal factor.
Febs Lett., 589, 2015
|
|
2QVY
 
 | | 4-Chlorobenzoyl-CoA Ligase/Synthetase, I303G mutation, bound to 3,4-Dichlorobenzoate | | Descriptor: | 3,4-dichlorobenzoate, 4-Chlorobenzoate CoA Ligase | | Authors: | Wu, R, Reger, A.S, Cao, J, Gulick, A.M, Dunaway-Mariano, D. | | Deposit date: | 2007-08-09 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Rational redesign of the 4-chlorobenzoate binding site of 4-chlorobenzoate: coenzyme a ligase for expanded substrate range.
Biochemistry, 46, 2007
|
|
4XM7
 
 | | Anthrax toxin lethal factor with ligand-induced binding pocket | | Descriptor: | 1,2-ETHANEDIOL, Lethal factor, N~2~-[(4-fluoro-3-methoxyphenyl)sulfonyl]-N-hydroxy-N~2~-(2-methylpropyl)-D-valinamide, ... | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2015-01-14 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ligand-induced expansion of the S1' site in the anthrax toxin lethal factor.
Febs Lett., 589, 2015
|
|
3V7S
 
 | | Crystal structure of Staphylococcus aureus biotin protein ligase in complex with inhibitor 0364 | | Descriptor: | 5-methyl-3-[4-(4-{5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentyl}-1H-1,2,3-triazol-1-yl)butyl]-1,3-benzoxazol-2(3H)-one, Biotin ligase | | Authors: | Yap, M.Y, Pendini, N.R. | | Deposit date: | 2011-12-21 | | Release date: | 2012-04-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Selective inhibition of biotin protein ligase from Staphylococcus aureus.
J.Biol.Chem., 287, 2012
|
|
2QHO
 
 | |
3V8K
 
 | |
2INX
 
 | | Crystal Structure of Ketosteroid Isomerase D40N from Pseudomonas putida (pKSI) with bound 2,6-difluorophenol | | Descriptor: | 2,6-DIFLUOROPHENOL, Steroid delta-isomerase | | Authors: | Martinez Caaveiro, J.M, Pybus, B, Ringe, D, Petsko, G.A, Sigala, P, Kraut, D, Herschlag, D. | | Deposit date: | 2006-10-09 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Testing geometrical discrimination within an enzyme active site: constrained hydrogen bonding in the ketosteroid isomerase oxyanion hole.
J.Am.Chem.Soc., 130, 2008
|
|
5WZZ
 
 | | The SIAH E3 ubiquitin ligases promote Wnt/ beta-catenin signaling through mediating Wnt-induced Axin degradation | | Descriptor: | Axin-1, E3 ubiquitin-protein ligase SIAH1, ZINC ION | | Authors: | Ji, L, Jiang, B, Jiang, X, Charlat, O, Chen, A, Mickanin, C, Bauer, A, Xu, W, Yan, X.-X, Cong, F. | | Deposit date: | 2017-01-19 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | The SIAH E3 ubiquitin ligases promote Wnt/ beta-catenin signaling through mediating Wnt-induced Axin degradation
Genes Dev., 31, 2017
|
|
3V7R
 
 | | Crystal structure of Staphylococcus aureus biotin protein ligase in complex with inhibitor | | Descriptor: | (3aS,4S,6aR)-4-(5-{1-[4-(6-amino-9H-purin-9-yl)butyl]-1H-1,2,3-triazol-4-yl}pentyl)tetrahydro-1H-thieno[3,4-d]imidazol-2(3H)-one, Biotin ligase | | Authors: | Yap, M.Y, Pendini, N.R. | | Deposit date: | 2011-12-21 | | Release date: | 2012-12-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Selective inhibition of biotin protein ligase from Staphylococcus aureus.
J.Biol.Chem., 287, 2012
|
|
3V8L
 
 | |
1FVI
 
 | | CRYSTAL STRUCTURE OF CHLORELLA VIRUS DNA LIGASE-ADENYLATE | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORELLA VIRUS DNA LIGASE-ADENYLATE, SULFATE ION | | Authors: | Odell, M, Sriskanda, V, Shuman, S, Nikolov, D.B. | | Deposit date: | 2000-09-20 | | Release date: | 2000-11-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of eukaryotic DNA ligase-adenylate illuminates the mechanism of nick sensing and strand joining.
Mol.Cell, 6, 2000
|
|
2QVX
 
 | | 4-Chlorobenzoyl-CoA Ligase/Synthetase, I303G mutation, bound to 3-Chlorobenzoate | | Descriptor: | 3-chlorobenzoate, 4-Chlorobenzoate CoA Ligase | | Authors: | Wu, R, Reger, A.S, Cao, J, Gulick, A.M, Dunaway-Mariano, D. | | Deposit date: | 2007-08-09 | | Release date: | 2007-12-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Rational redesign of the 4-chlorobenzoate binding site of 4-chlorobenzoate: coenzyme a ligase for expanded substrate range.
Biochemistry, 46, 2007
|
|
2QVZ
 
 | | 4-Chlorobenzoyl-CoA Ligase/Synthetase, I303A mutation, bound to 3-Chlorobenzoate | | Descriptor: | 3-chlorobenzoate, 4-Chlorobenzoate CoA Ligase/Synthetase | | Authors: | Wu, R, Reger, A.S, Cao, J, Gulick, A.M, Dunaway-Mariano, D. | | Deposit date: | 2007-08-09 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rational redesign of the 4-chlorobenzoate binding site of 4-chlorobenzoate: coenzyme a ligase for expanded substrate range.
Biochemistry, 46, 2007
|
|
2QW0
 
 | | 4-Chlorobenzoyl-CoA Ligase/Synthetase, I303A mutation, bound to 3,4 Dichlorobenzoate | | Descriptor: | 3,4-dichlorobenzoate, 4-Chlorobenzoate CoA Ligase | | Authors: | Wu, R, Reger, A.S, Cao, J, Gulick, A.M, Dunaway-Mariano, D. | | Deposit date: | 2007-08-09 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Rational redesign of the 4-chlorobenzoate binding site of 4-chlorobenzoate: coenzyme a ligase for expanded substrate range.
Biochemistry, 46, 2007
|
|
4ZQI
 
 | | Crystal structure of Apo D-alanine-D-alanine ligase(DDL) from Yersinia pestis | | Descriptor: | D-alanine--D-alanine ligase, SODIUM ION | | Authors: | Tran, H.-T, Kang, L.-W, Hong, M.-K, Ngo, H.P.T, Huynh, K.H, Ahn, Y.J. | | Deposit date: | 2015-05-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of D-alanine-D-alanine ligase from Yersinia pestis: nucleotide phosphate recognition by the serine loop.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
7M4O
 
 | |
7M4M
 
 | |
7M4N
 
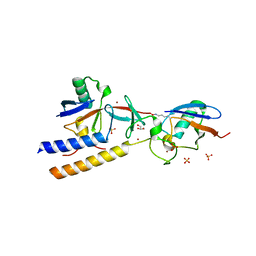 | |
5C1O
 
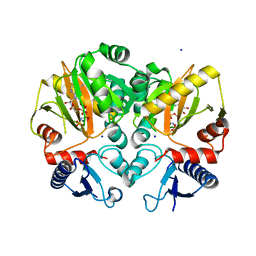 | | Crystal structure of AMP-PNP complexed D-alanine-D-alanine ligase(DDL) from Yersinia pestis | | Descriptor: | D-alanine--D-alanine ligase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Tran, H.T, Kang, L.W, Hong, M.K. | | Deposit date: | 2015-06-15 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of D-alanine-D-alanine ligase from Yersinia pestis: nucleotide phosphate recognition by the serine loop.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
7DBS
 
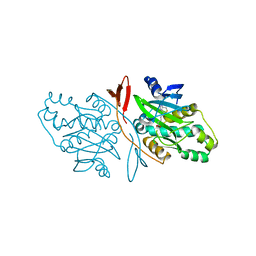 | |
3ATP
 
 | | Structure of the ligand binding domain of the bacterial serine chemoreceptor Tsr with ligand | | Descriptor: | Methyl-accepting chemotaxis protein I, SERINE | | Authors: | Tajima, H, Sakuma, M, Homma, K, Kawagishi, I, Imada, K. | | Deposit date: | 2011-01-07 | | Release date: | 2011-10-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand specificity determined by differentially arranged common ligand-binding residues in bacterial amino acid chemoreceptors Tsr and Tar.
J.Biol.Chem., 286, 2011
|
|
5C1P
 
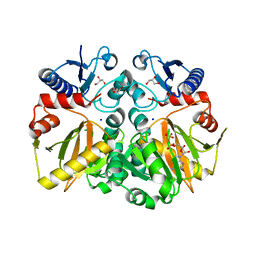 | | Crystal structure of ADP and D-alanyl-D-alanine complexed D-alanine-D-alanine ligase(DDL) from Yersinia pestis | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, D-ALANINE, ... | | Authors: | Tran, H.T, Kang, L.W, Hong, M.K, Ngo, H.P.T. | | Deposit date: | 2015-06-15 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of D-alanine-D-alanine ligase from Yersinia pestis: nucleotide phosphate recognition by the serine loop.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
