4K6I
 
 | | Crystal structure of human retinoid X receptor alpha-ligand binding domain complex with Targretin and the coactivator peptide GRIP-1 | | Descriptor: | 4-[1-(3,5,5,8,8-pentamethyl-5,6,7,8-tetrahydronaphthalen-2-yl)ethenyl]benzoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Xia, G, Smith, C.D, Muccio, D.D. | | Deposit date: | 2013-04-15 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Defining the Communication between Agonist and Coactivator Binding in the Retinoid X Receptor alpha Ligand Binding Domain.
J.Biol.Chem., 289, 2014
|
|
4AIP
 
 | | The FrpB iron transporter from Neisseria meningitidis (F3-3 variant) | | Descriptor: | 3,6,9,12,15-PENTAOXATRICOSAN-1-OL, FE-REGULATED PROTEIN B, GLUTAMIC ACID | | Authors: | Saleem, M, Prince, S.M, Derrick, J.P. | | Deposit date: | 2012-02-11 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Use of a Molecular Decoy to Segregate Transport from Antigenicity in the Frpb Iron Transporter from Neisseria Meningitidis
Plos One, 8, 2013
|
|
2UUV
 
 | | alkyldihydroxyacetonephosphate synthase in P1 | | Descriptor: | ALKYLDIHYDROXYACETONEPHOSPHATE SYNTHASE, FLAVIN-ADENINE DINUCLEOTIDE, HEXADECAN-1-OL | | Authors: | Razeto, A, Mattiroli, F, Carpanelli, E, Aliverti, A, Pandini, V, Coda, A, Mattevi, A. | | Deposit date: | 2007-03-07 | | Release date: | 2007-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The Crucial Step in Ether Phospholipid Biosynthesis: Structural Basis of a Noncanonical Reaction Associated with a Peroxisomal Disorder.
Structure, 15, 2007
|
|
8DIW
 
 | | Crystal structure of NavAb E96P as a basis for the human Nav1.7 Inherited Erythromelalgia S211P mutation | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Ion transport protein | | Authors: | Wisedchaisri, G, Gamal El-Din, T.M, Zheng, N, Catterall, W.A. | | Deposit date: | 2022-06-29 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural basis for severe pain caused by mutations in the voltage sensors of sodium channel NaV1.7.
J.Gen.Physiol., 155, 2023
|
|
6ENX
 
 | | Zebrafish Sirt5 in complex with stalled bicyclic intermediate of inhibitory compound 10 | | Descriptor: | 4-[(2~{R},3~{a}~{R},5~{R},6~{R},6~{a}~{R})-5-[[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxymethyl]-2-[[(5~{S})-6-[[(2~{S})-3-(1~{H}-indol-3-yl)-1-oxidanylidene-1-(propan-2-ylamino)propan-2-yl]amino]-6-oxidanylidene-5-(phenylmethoxycarbonylamino)hexyl]amino]-6-oxidanyl-3~{a},5,6,6~{a}-tetrahydrofuro[2,3-d][1,3]oxathiol-2-yl]butanoic acid, DIMETHYL SULFOXIDE, NAD-dependent protein deacylase sirtuin-5, ... | | Authors: | Pannek, M, Steegborn, C. | | Deposit date: | 2017-10-06 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mechanism-Based Inhibitors of the Human Sirtuin 5 Deacylase: Structure-Activity Relationship, Biostructural, and Kinetic Insight.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5N2J
 
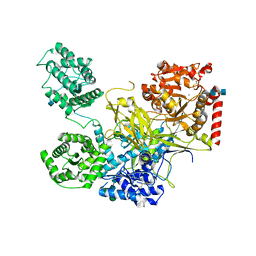 | | UDP-Glucose Glycoprotein Glucosyltransferase from Chaetomium thermophilum (closed form) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Roversi, P, Caputo, A.T, Hill, J, Alonzi, D.S, Zitzmann, N. | | Deposit date: | 2017-02-07 | | Release date: | 2017-07-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Interdomain conformational flexibility underpins the activity of UGGT, the eukaryotic glycoprotein secretion checkpoint.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8DIY
 
 | | Crystal structure of NavAb L101S as a basis for the human Nav1.7 Inherited Erythromelalgia F216S mutation | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Ion transport protein | | Authors: | Wisedchaisri, G, Gamal El-Din, T.M, Zheng, N, Catterall, W.A. | | Deposit date: | 2022-06-29 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis for severe pain caused by mutations in the voltage sensors of sodium channel NaV1.7.
J.Gen.Physiol., 155, 2023
|
|
3IF7
 
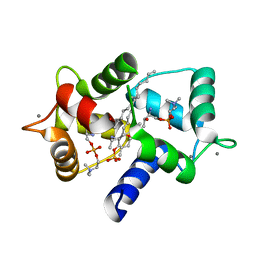 | | Structure of Calmodulin complexed with its first endogenous inhibitor, sphingosylphosphorylcholine | | Descriptor: | 2-{[(R)-{[(2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl]oxy}(hydroxy)phosphoryl]oxy}-N,N,N-trimethylethanaminium, CALCIUM ION, Calmodulin | | Authors: | Kovacs, E, Harmat, V, Toth, J, Vertessy, B.G, Modos, K, Kardos, J, Liliom, K. | | Deposit date: | 2009-07-24 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and mechanism of calmodulin binding to a signaling sphingolipid reveal new aspects of lipid-protein interactions
Faseb J., 24, 2010
|
|
1RBV
 
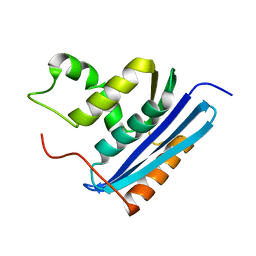 | | STRUCTURAL STUDY OF MUTANTS OF ESCHERICHIA COLI RIBONUCLEASE HI WITH ENHANCED THERMOSTABILITY | | Descriptor: | RIBONUCLEASE H | | Authors: | Ishikawa, K, Kimura, S, Kanaya, S, Morikawa, K, Nakamura, H. | | Deposit date: | 1993-02-16 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural study of mutants of Escherichia coli ribonuclease HI with enhanced thermostability.
Protein Eng., 6, 1993
|
|
4PE3
 
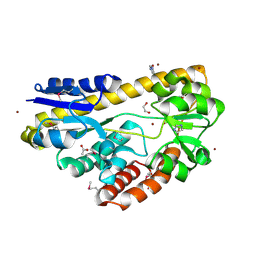 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM RHODOBACTER SPHAEROIDES (Rsph17029_3620, TARGET EFI-510199), APO OPEN STRUCTURE | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, IMIDAZOLE, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-22 | | Release date: | 2014-05-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
3IAP
 
 | | E. coli (lacZ) beta-galactosidase (E416Q) | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Lo, S, Dugdale, M.L, Jeerh, N, Ku, T, Roth, N.J, Huber, R.E. | | Deposit date: | 2009-07-14 | | Release date: | 2009-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Studies of Glu-416 variants of beta-galactosidase (E. coli) show that the active site Mg(2+) is not important for structure and indicate that the main role of Mg (2+) is to mediate optimization of active site chemistry
Protein J., 29, 2010
|
|
5D7D
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 7-propyl-3-[2-(pyridin-3-yl)-1,3-thiazol-5-yl]-1,7-dihydro-6H-pyrazolo[3,4-b]pyridin-6-one, CHLORIDE ION, ... | | Authors: | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | Deposit date: | 2015-08-13 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Indazole Derivatives as a Novel Class of Bacterial Gyrase B Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
2QYL
 
 | | Crystal structure of PDE4B2B in complex with inhibitor NPV | | Descriptor: | 4-[8-(3-nitrophenyl)-1,7-naphthyridin-6-yl]benzoic acid, MAGNESIUM ION, Phosphodiesterase 4B, ... | | Authors: | Ke, H. | | Deposit date: | 2007-08-15 | | Release date: | 2008-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the four subfamilies of phosphodiesterase-4 provide insight into the selectivity of their inhibitors.
Biochem.J., 408, 2007
|
|
4B33
 
 | | Humanised monomeric RadA in complex with napht-2-ol | | Descriptor: | 1-NAPHTHOL, DNA REPAIR AND RECOMBINATION PROTEIN RADA, PHOSPHATE ION | | Authors: | Scott, D.E, Ehebauer, M.T, Pukala, T, Marsh, M, Blundell, T.L, Venkitaraman, A.R, Abell, C, Hyvonen, M. | | Deposit date: | 2012-07-20 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Using a Fragment-Based Approach to Target Protein-Protein Interactions.
Chembiochem, 14, 2013
|
|
1WDV
 
 | | Crystal structure of hypothetical protein APE2540 | | Descriptor: | hypothetical protein APE2540 | | Authors: | Murayama, K, Kato-Murayama, M, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-18 | | Release date: | 2004-11-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a putative trans-editing enzyme for prolyl-tRNA synthetase from Aeropyrum pernix K1 at 1.7 A resolution.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
5TR6
 
 | | Discovery of TAK-659, an Orally Available Investigational Inhibitor of Spleen Tyrosine Kinase (SYK) | | Descriptor: | 1,2-ETHANEDIOL, 6-{[(1R,2S)-2-aminocyclohexyl]amino}-7-fluoro-4-(1-methyl-1H-pyrazol-4-yl)-1,2-dihydro-3H-pyrrolo[3,4-c]pyridin-3-one, Tyrosine-protein kinase SYK | | Authors: | Yano, J, Jennings, A, Lam, B, Hoffman, I.D. | | Deposit date: | 2016-10-25 | | Release date: | 2016-11-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Discovery of TAK-659 an orally available investigational inhibitor of Spleen Tyrosine Kinase (SYK).
Bioorg. Med. Chem. Lett., 26, 2016
|
|
5GQD
 
 | | Crystal structure of covalent glycosyl-enzyme intermediate of xylanase mutant (T82A, N127S, and E128H) from Streptomyces olivaceoviridis E-86 | | Descriptor: | Beta-xylanase, GLYCEROL, beta-D-xylopyranose-(1-4)-alpha-D-xylopyranose | | Authors: | Suzuki, R, Fujimoto, Z, Kaneko, S, Kuno, A. | | Deposit date: | 2016-08-07 | | Release date: | 2017-08-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Azidolysis by the Formation of Stable Ser-His Catalytic Dyad in a Glycoside Hydrolase Family 10 Xylanase Mutant
J.Appl.Glyosci., 65, 2019
|
|
3IE4
 
 | | b-glucan binding domain of Drosophila GNBP3 defines a novel family of pattern recognition receptor | | Descriptor: | 1,2-ETHANEDIOL, Gram-Negative Binding Protein 3, ZINC ION | | Authors: | Mishima, Y, Coste, F, Kellenberger, C, Roussel, A. | | Deposit date: | 2009-07-22 | | Release date: | 2009-08-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The N-terminal domain of drosophila gram-negative binding protein 3 (GNBP3) defines a novel family of fungal pattern recognition receptors
To be Published
|
|
3L2K
 
 | | Structure of phenazine antibiotic biosynthesis protein with substrate | | Descriptor: | EhpF, phenazine-1,6-dicarboxylic acid | | Authors: | Bera, A.K, Atanasova, V, Parsons, J.F. | | Deposit date: | 2009-12-15 | | Release date: | 2010-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the D-alanylgriseoluteic acid biosynthetic protein EhpF, an atypical member of the ANL superfamily of adenylating enzymes.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
5DAL
 
 | |
2ALC
 
 | | ETHANOL REGULON TRANSCRIPTIONAL ACTIVATOR DNA-BINDING DOMAIN FROM ASPERGILLUS NIDULANS | | Descriptor: | PROTEIN (ETHANOL REGULON TRANSCRIPTIONAL ACTIVATOR), ZINC ION | | Authors: | Cerdan, R, Cahuzac, B, Felenbok, B, Guittet, E. | | Deposit date: | 1999-01-20 | | Release date: | 2000-01-21 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of AlcR (1-60) provides insight in the unusual DNA binding properties of this zinc binuclear cluster protein.
J.Mol.Biol., 295, 2000
|
|
5JAT
 
 | |
3ILA
 
 | |
8E3L
 
 | | E. coli 50S ribosome bound to D-linker solithromycin conjugate | | Descriptor: | (2~{R})-~{N}-[(2~{R})-6-azanyl-1-[[(2~{R})-1-[[(2~{R})-1-[[3-[1-[4-[(1~{S},2~{R},5~{S},7~{R},8~{R},9~{R},11~{R},13~{R},14~{R})-8-[(2~{S},3~{R},4~{S},6~{R})-4-(dimethylamino)-6-methyl-3-oxidanyl-oxan-2-yl]oxy-2-ethyl-5-fluoranyl-9-methoxy-1,5,7,11,13-pentamethyl-4,6,12,16-tetrakis(oxidanylidene)-3,17-dioxa-15-azabicyclo[12.3.0]heptadecan-15-yl]butyl]-1,2,3-triazol-4-yl]phenyl]amino]-4-methylsulfanyl-1-oxidanylidene-butan-2-yl]amino]-3-(4-hydroxyphenyl)-1-oxidanylidene-propan-2-yl]amino]-1-oxidanylidene-hexan-2-yl]-1-[(2~{R})-2-[[(2~{R})-2-[2-[[2,3-bis(oxidanyl)phenyl]carbonyl-[4-[[2,3-bis(oxidanyl)phenyl]carbonylamino]butyl]amino]ethanoylamino]-3-(1~{H}-indol-3-yl)propanoyl]amino]-3-oxidanyl-propanoyl]pyrrolidine-2-carboxamide, 50S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2022-08-17 | | Release date: | 2023-06-28 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Solithromycin siderophore conjugates
To Be Published
|
|
7JQ1
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI4 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-L-valyl-N-{(2S)-1-hydroxy-3-[(3R)-2-oxo-3,4-dihydro-2H-pyrrol-3-yl]propan-2-yl}-L-phenylalaninamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Quick Route to Multiple Highly Potent SARS-CoV-2 Main Protease Inhibitors*.
Chemmedchem, 16, 2021
|
|
