3HHI
 
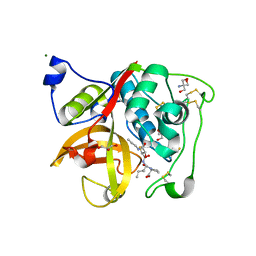 | | Crystal Structure of Cathepsin B from T. brucei in complex with CA074 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cathepsin B-like cysteine protease, GLYCEROL, ... | | Authors: | Wu, P, Kerr, I.D, Brinen, L.S. | | Deposit date: | 2009-05-15 | | Release date: | 2010-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of TbCatB and rhodesain, potential chemotherapeutic targets and major cysteine proteases of Trypanosoma brucei
Plos Negl Trop Dis, 4, 2010
|
|
2JHL
 
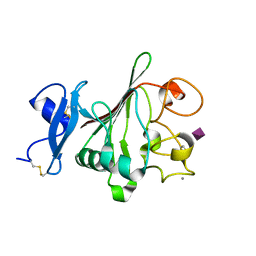 | | Structure of globular heads of M-ficolin complexed with sialic acid | | Descriptor: | CALCIUM ION, FICOLIN-1, N-acetyl-beta-neuraminic acid | | Authors: | Garlatti, V, Martin, L, Gout, E, Reiser, J.B, Arlaud, G.J, Thielens, N.M, Gaboriaud, C. | | Deposit date: | 2007-02-22 | | Release date: | 2007-10-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Innate Immune Sensing by M-Ficolin and its Control by a Ph-Dependent Conformational Switch.
J.Biol.Chem., 282, 2007
|
|
256D
 
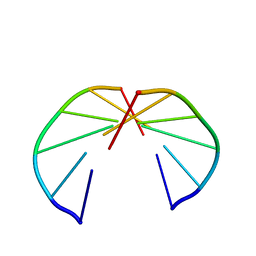 | |
3HEI
 
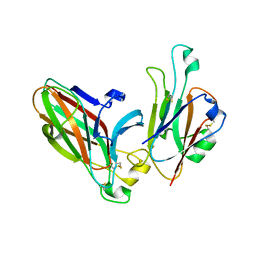 | | Ligand Recognition by A-Class Eph Receptors: Crystal Structures of the EphA2 Ligand-Binding Domain and the EphA2/ephrin-A1 Complex | | Descriptor: | Ephrin type-A receptor 2, Ephrin-A1 | | Authors: | Himanen, J.P, Goldgur, Y, Miao, H, Myshkin, E, Guo, H, Buck, M, Nguyen, M, Rajashankar, K.R, Wang, B, Nikolov, D.B. | | Deposit date: | 2009-05-08 | | Release date: | 2009-06-30 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ligand recognition by A-class Eph receptors: crystal structures of the EphA2 ligand-binding domain and the EphA2/ephrin-A1 complex.
Embo Rep., 10, 2009
|
|
1EKW
 
 | | NMR STRUCTURE OF A DNA THREE-WAY JUNCTION | | Descriptor: | DNA (5'-D(*CP*GP*GP*TP*GP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*CP*CP*AP*CP*CP*G)-3'), DNA (5'-D(*GP*GP*AP*CP*GP*TP*CP*GP*CP*AP*GP*C)-3') | | Authors: | Thiviyanathan, V, Luxon, B.A, Leontis, N.B, Donne, D, Gorenstein, D.G. | | Deposit date: | 2000-03-09 | | Release date: | 2000-03-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Hybrid-hybrid matrix structural refinement of a DNA three-way junction from 3D NOESY-NOESY.
J.Biomol.NMR, 14, 1999
|
|
2ANL
 
 | | X-ray crystal structure of the aspartic protease plasmepsin 4 from the malarial parasite plasmodium malariae bound to an allophenylnorstatine based inhibitor | | Descriptor: | (4R)-3-{(2S,3S)-2-hydroxy-3-[(3-hydroxy-2-methylbenzoyl)amino]-4-phenylbutanoyl}-5,5-dimethyl-N-(2-methylbenzyl)-1,3-thiazolidine-4-carboxamide, plasmepsin IV | | Authors: | Clemente, J.C, Govindasamy, L, Madabushi, A, Fisher, S.Z, Moose, R.E, Yowell, C.A, Hidaka, K, Kimura, T, Hayashi, Y, Kiso, Y, Agbandje-McKenna, M, Dame, J.B, Dunn, B.M, McKenna, R. | | Deposit date: | 2005-08-11 | | Release date: | 2006-04-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the aspartic protease plasmepsin 4 from the malarial parasite Plasmodium malariae bound to an allophenylnorstatine-based inhibitor.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2AMV
 
 | | THE STRUCTURE OF GLYCOGEN PHOSPHORYLASE B WITH AN ALKYL-DIHYDROPYRIDINE-DICARBOXYLIC ACID | | Descriptor: | 2,3-DICARBOXY-4-(2-CHLORO-PHENYL)-1-ETHYL-5-ISOPROPOXYCARBONYL-6-METHYL-PYRIDINIUM, GLYCEROL, PROTEIN (GLYCOGEN PHOSPHORYLASE), ... | | Authors: | Zographos, S.E, Oikonomakos, N.G, Johnson, L.N. | | Deposit date: | 1998-10-13 | | Release date: | 1998-10-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of glycogen phosphorylase b with an alkyldihydropyridine-dicarboxylic acid compound, a novel and potent inhibitor.
Structure, 5, 1997
|
|
1HSW
 
 | |
5PAL
 
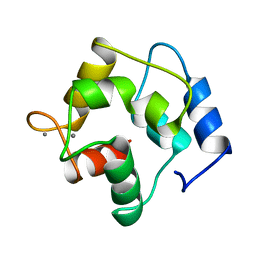 | | CRYSTAL STRUCTURE OF THE UNIQUE PARVALBUMIN COMPONENT FROM MUSCLE OF THE LEOPARD SHARK (TRIAKIS SEMIFASCIATA). THE FIRST X-RAY STUDY OF AN ALPHA-PARVALBUMIN | | Descriptor: | CALCIUM ION, PARVALBUMIN | | Authors: | Roquet, F, Declercq, J.-P, Tinant, B, Rambaud, J, Parello, J. | | Deposit date: | 1991-09-25 | | Release date: | 1993-10-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structure of the unique parvalbumin component from muscle of the leopard shark (Triakis semifasciata). The first X-ray study of an alpha-parvalbumin.
J.Mol.Biol., 223, 1992
|
|
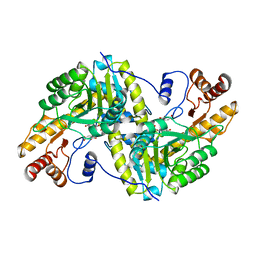
2CST
 
 | |
4K4S
 
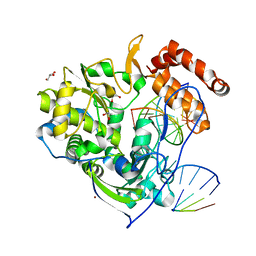 | | Poliovirus polymerase elongation complex (r3_form) | | Descriptor: | GLYCEROL, RNA (5'-R(*AP*AP*GP*UP*CP*UP*CP*CP*AP*GP*GP*UP*CP*UP*CP*UP*CP*GP*CP*GP*AP*AP*A)-3'), RNA (5'-R(*GP*GP*GP*AP*GP*AP*UP*GP*A)-3'), ... | | Authors: | Gong, P, Peersen, O.B. | | Deposit date: | 2013-04-12 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of coxsackievirus, rhinovirus, and poliovirus polymerase elongation complexes solved by engineering RNA mediated crystal contacts.
Plos One, 8, 2013
|
|
154L
 
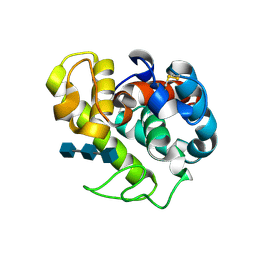 | |
1SNU
 
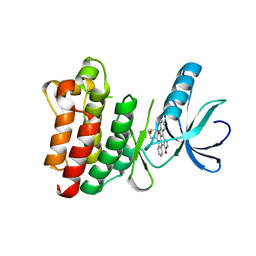 | | CRYSTAL STRUCTURE OF THE UNPHOSPHORYLATED INTERLEUKIN-2 TYROSINE KINASE CATALYTIC DOMAIN | | Descriptor: | STAUROSPORINE, Tyrosine-protein kinase ITK/TSK | | Authors: | Brown, K, Long, J.M, Vial, S.C, Dedi, N, Dunster, N.J, Renwick, S.B, Tanner, A.J, Frantz, J.D, Fleming, M.A, Cheetham, G.M.T. | | Deposit date: | 2004-03-12 | | Release date: | 2004-07-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of interleukin-2 tyrosine kinase and their implications for the design of selective inhibitors.
J.Biol.Chem., 279, 2004
|
|
1SBB
 
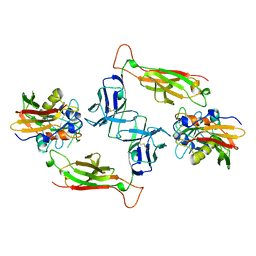 | | T-CELL RECEPTOR BETA CHAIN COMPLEXED WITH SUPERANTIGEN SEB | | Descriptor: | PROTEIN (14.3.D T CELL ANTIGEN RECEPTOR), PROTEIN (STAPHYLOCOCCAL ENTEROTOXIN B) | | Authors: | Li, H, Mariuzza, R.A. | | Deposit date: | 1999-02-22 | | Release date: | 1999-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Three-dimensional structure of the complex between a T cell receptor beta chain and the superantigen staphylococcal enterotoxin B.
Immunity, 9, 1998
|
|
1SGV
 
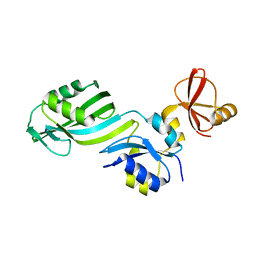 | | STRUCTURE OF TRNA PSI55 PSEUDOURIDINE SYNTHASE (TRUB) | | Descriptor: | tRNA pseudouridine synthase B | | Authors: | Chaudhuri, B.N, Chan, S, Perry, L.J, Yeates, T.O, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-02-24 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the apo forms of psi 55 tRNA pseudouridine synthase from Mycobacterium tuberculosis: a hinge at the base of the catalytic cleft.
J.Biol.Chem., 279, 2004
|
|
2IFW
 
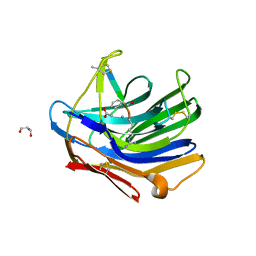 | | Crystal structure of scytalido-glutamic peptidase with a transition state analog inhibitor | | Descriptor: | ACETIC ACID, GLYCEROL, Heptapeptide, ... | | Authors: | Pillai, B, Cherney, M.M, Hiraga, K, Takada, K, Oda, K, James, M.N. | | Deposit date: | 2006-09-21 | | Release date: | 2006-10-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of scytalidoglutamic peptidase with its first potent inhibitor provides insights into substrate specificity and catalysis.
J.Mol.Biol., 365, 2007
|
|
1SM2
 
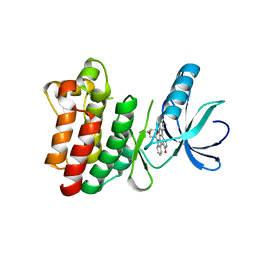 | | Crystal structure of the phosphorylated Interleukin-2 tyrosine kinase catalytic domain | | Descriptor: | STAUROSPORINE, Tyrosine-protein kinase ITK/TSK | | Authors: | Brown, K, Long, J.M, Vial, S.C.M, Dedi, N, Dunster, N.J, Renwick, S.B, Tanner, A.J, Frantz, J.D, Fleming, M.A, Cheetham, G.M.T. | | Deposit date: | 2004-03-08 | | Release date: | 2004-07-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of interleukin-2 tyrosine kinase and their implications for the design of selective inhibitors.
J.Biol.Chem., 279, 2004
|
|
4OOS
 
 | |
2MYZ
 
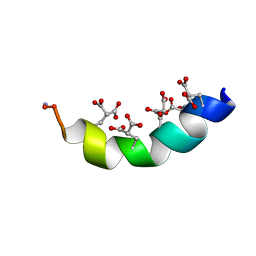 | | The Solution Structure of the Magnesium-bound Conantokin-R1B Mutant | | Descriptor: | Conantokin-R1-B | | Authors: | Kunda, S, Yuan, Y, Balsara, R.D, Zajicek, J, Castellino, F.J. | | Deposit date: | 2015-02-04 | | Release date: | 2015-06-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Hydroxyproline-induced Helical Disruption in Conantokin Rl-B Affects Subunit-selective Antagonistic Activities toward Ion Channels of N-Methyl-d-aspartate Receptors.
J.Biol.Chem., 290, 2015
|
|
2IRT
 
 | |
2MZM
 
 | | The Solution Structure of the Magnesium-bound Conantokin-G | | Descriptor: | Conantokin-R1-B | | Authors: | Kunda, S, Yuan, Y, Balsara, R.D, Zajicek, J, Castellino, F.J. | | Deposit date: | 2015-02-14 | | Release date: | 2015-06-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Hydroxyproline-induced Helical Disruption in Conantokin Rl-B Affects Subunit-selective Antagonistic Activities toward Ion Channels of N-Methyl-d-aspartate Receptors.
J.Biol.Chem., 290, 2015
|
|
2MZL
 
 | | The Solution Structure of the Magnesium-bound Conantokin-G Mutant | | Descriptor: | Conantokin-R1-B | | Authors: | Kunda, S, Yuan, Y, Balsara, R.D, Zajicek, J, Castellino, F.J. | | Deposit date: | 2015-02-14 | | Release date: | 2015-06-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Hydroxyproline-induced Helical Disruption in Conantokin Rl-B Affects Subunit-selective Antagonistic Activities toward Ion Channels of N-Methyl-d-aspartate Receptors.
J.Biol.Chem., 290, 2015
|
|
1B9U
 
 | |
1SVC
 
 | | NFKB P50 HOMODIMER BOUND TO DNA | | Descriptor: | DNA (5'-D(*AP*GP*AP*TP*GP*GP*GP*GP*AP*AP*TP*CP*CP*CP*CP*TP*A P*GP*A)-3'), PROTEIN (NUCLEAR FACTOR KAPPA-B (NF-KB)) | | Authors: | Mueller, C.W, Harrison, S.C. | | Deposit date: | 1995-11-27 | | Release date: | 1996-06-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the NF-kappa B p50 homodimer bound to DNA.
Nature, 373, 1995
|
|
2RFK
 
 | | Substrate RNA Positioning in the Archaeal H/ACA Ribonucleoprotein Complex | | Descriptor: | Probable tRNA pseudouridine synthase B, Ribosome biogenesis protein Nop10, Small nucleolar rnp similar to gar1, ... | | Authors: | Liang, B, Xue, S, Terns, R.M, Terns, M.P, Li, H, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2007-09-30 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Substrate RNA positioning in the archaeal H/ACA ribonucleoprotein complex.
Nat.Struct.Mol.Biol., 14, 2007
|
|
