1BHU
 
 | | THE 3D STRUCTURE OF THE STREPTOMYCES METALLOPROTEINASE INHIBITOR, SMPI, ISOLATED FROM STREPTOMYCES NIGRESCENS TK-23, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | METALLOPROTEINASE INHIBITOR | | Authors: | Tate, S, Ohno, A, Seeram, S.S, Hiraga, K, Oda, K, Kainosho, M. | | Deposit date: | 1998-06-10 | | Release date: | 1999-01-06 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the Streptomyces metalloproteinase inhibitor, SMPI, isolated from Streptomyces nigrescens TK-23: another example of an ancestral beta gamma-crystallin precursor structure.
J.Mol.Biol., 282, 1998
|
|
1F53
 
 | | NMR STRUCTURE OF KILLER TOXIN-LIKE PROTEIN SKLP | | Descriptor: | YEAST KILLER TOXIN-LIKE PROTEIN | | Authors: | Ohki, S, Kariya, E, Hiraga, K, Wakamiya, A, Isobe, T, Oda, K, Kainosho, M. | | Deposit date: | 2000-06-12 | | Release date: | 2000-12-27 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | NMR structure of Streptomyces killer toxin-like protein, SKLP: further evidence for the wide distribution of single-domain betagamma-crystallin superfamily proteins.
J.Mol.Biol., 305, 2001
|
|
5H6L
 
 | | DNA targeting ADP-ribosyltransferase Pierisin-1 in complex with beta-NAD+ | | Descriptor: | 1,2-ETHANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Pierisin-1 | | Authors: | Oda, T, Hirabayashi, H, Shikauchi, G, Takamura, R, Hiraga, K, Minami, H, Hashimoto, H, Yamamoto, M, Wakabayashi, K, Sugimura, T, Shimizu, T, Sato, M. | | Deposit date: | 2016-11-14 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of autoinhibition and activation of the DNA-targeting ADP-ribosyltransferase pierisin-1
J. Biol. Chem., 292, 2017
|
|
5H6M
 
 | | DNA targeting ADP-ribosyltransferase Pierisin-1 | | Descriptor: | 1,2-ETHANEDIOL, Pierisin-1 | | Authors: | Oda, T, Hirabayashi, H, Shikauchi, G, Takamura, R, Hiraga, K, Minami, H, Hashimoto, H, Yamamoto, M, Wakabayashi, K, Sugimura, T, Shimizu, T, Sato, M. | | Deposit date: | 2016-11-14 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of autoinhibition and activation of the DNA-targeting ADP-ribosyltransferase pierisin-1
J. Biol. Chem., 292, 2017
|
|
5H6K
 
 | | DNA targeting ADP-ribosyltransferase Pierisin-1 | | Descriptor: | 1,2-ETHANEDIOL, Pierisin-1 | | Authors: | Oda, T, Hirabayashi, H, Shikauchi, G, Takamura, R, Hiraga, K, Minami, H, Hashimoto, H, Yamamoto, M, Wakabayashi, K, Sugimura, T, Shimizu, T, Sato, M. | | Deposit date: | 2016-11-14 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of autoinhibition and activation of the DNA-targeting ADP-ribosyltransferase pierisin-1
J. Biol. Chem., 292, 2017
|
|
5H6N
 
 | | DNA targeting ADP-ribosyltransferase Pierisin-1, autoinhibitory form | | Descriptor: | Pierisin-1 | | Authors: | Oda, T, Hirabayashi, H, Shikauchi, G, Takamura, R, Hiraga, K, Minami, H, Hashimoto, H, Yamamoto, M, Wakabayashi, K, Sugimura, T, Shimizu, T, Sato, M. | | Deposit date: | 2016-11-14 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of autoinhibition and activation of the DNA-targeting ADP-ribosyltransferase pierisin-1
J. Biol. Chem., 292, 2017
|
|
5H6J
 
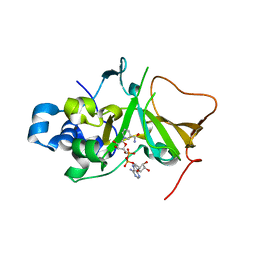 | | DNA targeting ADP-ribosyltransferase Pierisin-1 in complex with beta-NAD+ | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Pierisin-1 | | Authors: | Oda, T, Hirabayashi, H, Shikauchi, G, Takamura, R, Hiraga, K, Minami, H, Hashimoto, H, Yamamoto, M, Wakabayashi, K, Sugimura, T, Shimizu, T, Sato, M. | | Deposit date: | 2016-11-14 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of autoinhibition and activation of the DNA-targeting ADP-ribosyltransferase pierisin-1
J. Biol. Chem., 292, 2017
|
|
2IFW
 
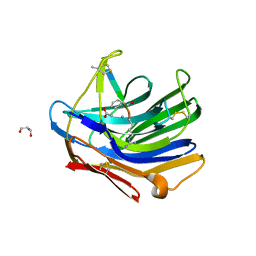 | | Crystal structure of scytalido-glutamic peptidase with a transition state analog inhibitor | | Descriptor: | ACETIC ACID, GLYCEROL, Heptapeptide, ... | | Authors: | Pillai, B, Cherney, M.M, Hiraga, K, Takada, K, Oda, K, James, M.N. | | Deposit date: | 2006-09-21 | | Release date: | 2006-10-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of scytalidoglutamic peptidase with its first potent inhibitor provides insights into substrate specificity and catalysis.
J.Mol.Biol., 365, 2007
|
|
2IFR
 
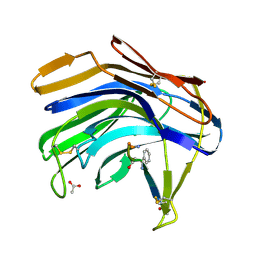 | | Crystal structure of Scytalido-glutamic peptidase with a peptide based transition state analog | | Descriptor: | ACETIC ACID, Octapeptide, Scytalidopepsin B | | Authors: | Pillai, B, Cherney, M.M, Hiraga, K, Takada, K, Oda, K, James, M.N. | | Deposit date: | 2006-09-21 | | Release date: | 2006-10-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of scytalidoglutamic peptidase with its first potent inhibitor provides insights into substrate specificity and catalysis.
J.Mol.Biol., 365, 2007
|
|
