7YN3
 
 | | Crystal structure of CcbD complex with CcbZ carrier protein domain | | Descriptor: | 1,1'-ethane-1,2-diyldipyrrolidine-2,5-dione, 4'-PHOSPHOPANTETHEINE, CcbD, ... | | Authors: | Mori, T, Lyu, S, Abe, I. | | Deposit date: | 2022-07-29 | | Release date: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of CcbD complex with carrier protein
To Be Published
|
|
6V8C
 
 | | Design, Synthesis, and Mechanism of Fluorine-substituted Cyclohexene Analogues of GAMA-Aminobutyric Acid (GABA) as Selective Ornithine Aminotransferase Inactivators | | Descriptor: | 3-aminocyclohexa-1,3-diene-1-carboxylic acid, Ornithine aminotransferase, mitochondrial, ... | | Authors: | Zhu, W, Doubleday, P.T, Catlin, D.S, Weerawarna, P, Butrin, A, Shen, S, Kelleher, N.L, Liu, D, Silverman, R.B. | | Deposit date: | 2019-12-10 | | Release date: | 2020-12-16 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design, Synthesis, and Mechanism of Fluorine-substituted Cyclohexene Analogues of GAMA-Aminobutyric Acid (GABA) as Selective Ornithine Aminotransferase Inactivators
To Be Published
|
|
2PKH
 
 | | Structural Genomics, the crystal structure of the C-terminal domain of histidine utilization repressor from Pseudomonas syringae pv. tomato str. DC3000 | | Descriptor: | 1,2-ETHANEDIOL, Histidine utilization repressor | | Authors: | Tan, K, Zhou, M, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-04-17 | | Release date: | 2007-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of the C-terminal domain of histidine utilization repressor from Pseudomonas syringae pv. tomato str. DC3000.
To be Published
|
|
3EN5
 
 | | Targeted polypharmacology: crystal structure of the c-Src kinase domain in complex with PP494, a multitargeted kinase inhibitor | | Descriptor: | 1-cyclobutyl-3-(3,4-dimethoxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Blair, J.A, Apsel, B, Knight, Z.A, Shokat, K.M. | | Deposit date: | 2008-09-25 | | Release date: | 2008-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Targeted polypharmacology: discovery of dual inhibitors of tyrosine and phosphoinositide kinases.
Nat.Chem.Biol., 4, 2008
|
|
6C1C
 
 | | FGFR1 kinase complex with inhibitor SN37116 | | Descriptor: | 7-(cyclohexylamino)-3-(2,6-dichloro-3,5-dimethoxyphenyl)-1-{(3S)-1-[(2E)-4-(dimethylamino)but-2-enoyl]pyrrolidin-3-yl}-3,4-dihydropyrimido[4,5-d]pyrimidin-2(1H)-one, Fibroblast growth factor receptor 1, SULFATE ION | | Authors: | Yosaatmadja, Y, Smaill, J.B, Squire, C.J. | | Deposit date: | 2018-01-04 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Understanding the structural requirements for covalent inhibition of FGFR1-3
To Be Published
|
|
6UYB
 
 | | Crystal structure of TEAD2 bound to Compound 1 | | Descriptor: | (3R,4R)-1-{3-[(E)-2-(4-chlorophenyl)ethenyl]-4-methoxy-5-methylphenyl}-3,4-dihydroxypyrrolidin-2-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ... | | Authors: | Noland, C.L, Holden, J.K, Crawford, J.J, Zbieg, J.R, Cunningham, C.N. | | Deposit date: | 2019-11-12 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.543 Å) | | Cite: | Small Molecule Dysregulation of TEAD Lipidation Induces a Dominant-Negative Inhibition of Hippo Pathway Signaling.
Cell Rep, 31, 2020
|
|
3EC7
 
 | | Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETIC ACID, ... | | Authors: | Kim, Y, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-08-29 | | Release date: | 2008-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Putative Dehydrogenase from Salmonella typhimurium LT2
To be Published
|
|
5BWM
 
 | | The complex structure of C3cer exoenzyme and GDP bound RhoA (NADH-bound state) | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ADP-ribosyltransferase, ... | | Authors: | Toda, A, Tsurumura, T, Yoshida, T, Tsuge, H. | | Deposit date: | 2015-06-08 | | Release date: | 2015-06-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rho GTPase Recognition by C3 Exoenzyme Based on C3-RhoA Complex Structure.
J.Biol.Chem., 290, 2015
|
|
5AR3
 
 | | RIP2 Kinase Catalytic Domain (1 - 310) complex with AMP-PCP | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, RECEPTOR-INTERACTING SERINE/THREONINE-PROTEIN KINASE 2 | | Authors: | Charnley, A.K, Convery, M.A, Lakdawala Shah, A, Jones, E, Hardwicke, P, Bridges, A, Votta, B.J, Gough, P.J, Marquis, R.W, Bertin, J, Casillas, L. | | Deposit date: | 2015-09-23 | | Release date: | 2015-10-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Crystal Structures of Human Rip2 Kinase Catalytic Domain Complexed with ATP-Competitive Inhibitors: Foundations for Understanding Inhibitor Selectivity.
Bioorg.Med.Chem., 23, 2015
|
|
6C9F
 
 | | AMP-activated protein kinase bound to pharmacological activator R734 | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1,5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Yan, Y, Zhou, X.E, Novick, S, Shaw, S.J, Li, Y, Hitoshi, Y, Brunzelle, J.S, Griffin, P.R, Xu, H.E, Melcher, K. | | Deposit date: | 2018-01-26 | | Release date: | 2018-11-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.924 Å) | | Cite: | Structures of AMP-activated protein kinase bound to novel pharmacological activators in phosphorylated, non-phosphorylated, and nucleotide-free states.
J. Biol. Chem., 294, 2019
|
|
3E3U
 
 | | Crystal structure of Mycobacterium tuberculosis peptide deformylase in complex with inhibitor | | Descriptor: | N-[(2R)-2-{[(2S)-2-(1,3-benzoxazol-2-yl)pyrrolidin-1-yl]carbonyl}hexyl]-N-hydroxyformamide, NICKEL (II) ION, Peptide deformylase | | Authors: | Meng, W, Xu, M, Pan, S, Koehn, J. | | Deposit date: | 2008-08-08 | | Release date: | 2009-01-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Peptide deformylase inhibitors of Mycobacterium tuberculosis: synthesis, structural investigations, and biological results.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
5Z5O
 
 | | Structure of Pycnonodysostosis disease related I249T mutant of human cathepsin K | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Biswas, S, Roy, S. | | Deposit date: | 2018-01-19 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Not all pycnodysostosis-related mutants of human cathepsin K are inactive - crystal structure and biochemical studies of an active mutant I249T.
FEBS J., 285, 2018
|
|
2Q88
 
 | | Crystal structure of EhuB in complex with ectoine | | Descriptor: | (4S)-2-METHYL-1,4,5,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, CADMIUM ION, Putative ABC transporter amino acid-binding protein | | Authors: | Hanekop, N, Hoeing, M, Sohn-Bosser, L, Jebbar, M, Schmitt, L, Bremer, E. | | Deposit date: | 2007-06-09 | | Release date: | 2008-01-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the ligand-binding protein EhuB from Sinorhizobium meliloti reveals substrate recognition of the compatible solutes ectoine and hydroxyectoine.
J.Mol.Biol., 374, 2007
|
|
4AQL
 
 | | HUMAN GUANINE DEAMINASE IN COMPLEX WITH VALACYCLOVIR | | Descriptor: | 2-[(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)methoxy]ethyl L-valinate, GUANINE DEAMINASE, ZINC ION | | Authors: | Welin, M, Egeblad, L, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kotenyova, T, Moche, M, Nyman, T, Persson, C, Schuler, H, Thorsell, A.G, Tresaugues, L, Weigelt, J, Nordlund, P. | | Deposit date: | 2012-04-18 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Pan-Pathway Based Interaction Profiling of Fda-Approved Nucleoside and Nucleobase Analogs with Enzymes of the Human Nucleotide Metabolism.
Plos One, 7, 2012
|
|
2X4Z
 
 | | Crystal Structure of the Human p21-Activated Kinase 4 in Complex with PF-03758309 | | Descriptor: | GLYCEROL, PF-3758309, SERINE/THREONINE-PROTEIN KINASE PAK 4 | | Authors: | Knighton, D.R, Deng, Y, Murray, B, Guo, C, Piraino, J, Westwick, J, Zhang, C, Lamerdin, J, Dagostino, E, Loi, C.-M, Zager, M, Kraynov, E, Christensen, J, Martinez, R, Kephart, S, Marakovits, J, Karlicek, S, Bergqvist, S, Smeal, T. | | Deposit date: | 2010-02-03 | | Release date: | 2010-05-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Small-Molecule P21-Activated Kinase Inhibitor Pf- 3758309 is a Potent Inhibitor of Oncogenic Signaling and Tumor Growth.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7GEZ
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ALP-POS-6747fa38-1 (Mpro-x11541) | | Descriptor: | 2-(4-acetylpiperazin-1-yl)-N-(isoquinolin-4-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
5C27
 
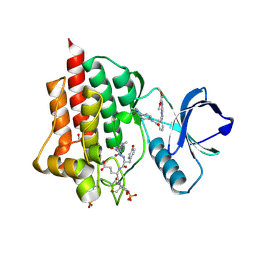 | | Crystal structure of SYK in complex with compound 2 | | Descriptor: | 3-{8-[(3,4-dimethoxyphenyl)amino]imidazo[1,2-a]pyrazin-6-yl}-N-[4-(methylcarbamoyl)phenyl]benzamide, GLU-VAL-TYR-GLU-SER, GLYCEROL, ... | | Authors: | Han, S, Chang, J. | | Deposit date: | 2015-06-15 | | Release date: | 2015-10-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Imidazotriazines: Spleen Tyrosine Kinase (Syk) Inhibitors Identified by Free-Energy Perturbation (FEP).
Chemmedchem, 11, 2016
|
|
5C7C
 
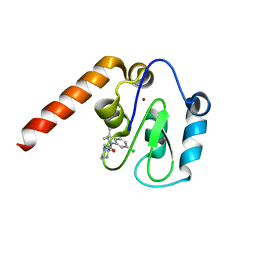 | | Fragment-Based Drug Discovery Targeting Inhibitor of Apoptosis Proteins: Compound 18 | | Descriptor: | (2R)-4-[2-(6-chloro-3,3-dimethyl-2,3-dihydro-1H-indol-1-yl)-2-oxoethyl]-2-methylpiperazin-1-ium, E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Chessari, G, Buck, I.M, Day, J.E.H, Day, P.J, Iqbal, A, Johnson, C.N, Lewis, E.J, Martins, V, Miller, D, Reader, M, Rees, D.C, Rich, S.J, Tamanini, E, Vitorino, M, Ward, G.A, Williams, P.A, Williams, G, Wilsher, N.E, Woolford, A.J.-A. | | Deposit date: | 2015-06-24 | | Release date: | 2015-08-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Fragment-Based Drug Discovery Targeting Inhibitor of Apoptosis Proteins: Discovery of a Non-Alanine Lead Series with Dual Activity Against cIAP1 and XIAP.
J.Med.Chem., 58, 2015
|
|
3G9J
 
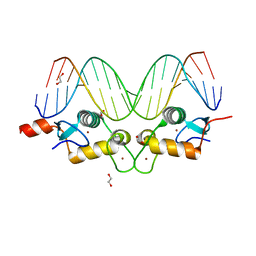 | | GR DNA binding domain:Pal, 18bp complex-36 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*GP*AP*AP*CP*AP*AP*AP*AP*TP*GP*TP*TP*CP*TP*G)-3'), DNA (5'-D(*CP*CP*AP*GP*AP*AP*CP*AP*TP*TP*TP*TP*GP*TP*TP*CP*TP*G)-3'), ... | | Authors: | Pufall, M.A, Yamamoto, K.R, Meijsing, S.H. | | Deposit date: | 2009-02-13 | | Release date: | 2009-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | DNA binding site sequence directs glucocorticoid receptor structure and activity.
Science, 324, 2009
|
|
4AD9
 
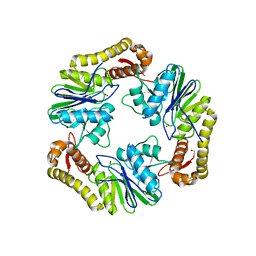 | | Crystal structure of human LACTB2. | | Descriptor: | 1,2-ETHANEDIOL, BETA-LACTAMASE-LIKE PROTEIN 2, ZINC ION | | Authors: | Allerston, C.K, Krojer, T, Shrestha, B, Burgess Brown, N, Chalk, R, Elkins, J.M, Filippakopoulos, P, Pike, A.C.W, Muniz, J.R.C, Vollmar, M, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, von Delft, F, Gileadi, O. | | Deposit date: | 2011-12-22 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of Lactb2, a Metallo-Beta-Lactamase Protein, as a Human Mitochondrial Endoribonuclease
Nucleic Acids Res., 44, 2016
|
|
4KWA
 
 | | Crystal structure of a putative transcriptional regulator from Saccharomonospora viridis in complex with choline | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, CHOLINE ION, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Endres, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-23 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a putative transcriptional regulator from Saccharomonospora viridis in complex with choline
To be Published
|
|
6MX8
 
 | | Crystal structure of anaplastic lymphoma kinase (ALK) bound by Brigatinib | | Descriptor: | 5-chloro-N~4~-[2-(dimethylphosphoryl)phenyl]-N~2~-{2-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl}pyrimidine-2,4-diamine, ALK tyrosine kinase receptor | | Authors: | Dougan, D.R, Zhou, T. | | Deposit date: | 2018-10-30 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Discovery of Brigatinib (AP26113), a Phosphine Oxide-Containing, Potent, Orally Active Inhibitor of Anaplastic Lymphoma Kinase.
J. Med. Chem., 59, 2016
|
|
5C3S
 
 | | Crystal structure of the full-length Neurospora crassa T7H in complex with alpha-KG and 5-formyluracil (5fU) | | Descriptor: | 1,2-ETHANEDIOL, 2,4-dioxo-1,2,3,4-tetrahydropyrimidine-5-carbaldehyde, 2-OXOGLUTARIC ACID, ... | | Authors: | Li, W, Zhang, T, Ding, J. | | Deposit date: | 2015-06-17 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular basis for the substrate specificity and catalytic mechanism of thymine-7-hydroxylase in fungi
Nucleic Acids Res., 43, 2015
|
|
5LSQ
 
 | | Ethylene Forming Enzyme from Pseudomonas syringae pv. phaseolicola - I222 crystal form | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Ethylene Forming Enzyme, ... | | Authors: | Zhang, Z, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2016-09-05 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and stereoelectronic insights into oxygenase-catalyzed formation of ethylene from 2-oxoglutarate.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5CBE
 
 | | E10 in complex with CXCL13 | | Descriptor: | 1,2-ETHANEDIOL, C-X-C motif chemokine 13, E10 heavy chain, ... | | Authors: | Tu, C, Bard, J, Mosyak, L. | | Deposit date: | 2015-06-30 | | Release date: | 2015-11-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Combination of Structural and Empirical Analyses Delineates the Key Contacts Mediating Stability and Affinity Increases in an Optimized Biotherapeutic Single-chain Fv (scFv).
J. Biol. Chem., 291, 2016
|
|
