4ZAM
 
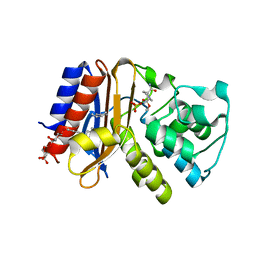 | | Crystal structure of SHV-1 beta-lactamase bound to avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase SHV-1, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE | | Authors: | Krishnan, N, van den Akker, F. | | Deposit date: | 2015-04-13 | | Release date: | 2016-01-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Inhibition of Klebsiella beta-Lactamases (SHV-1 and KPC-2) by Avibactam: A Structural Study.
Plos One, 10, 2015
|
|
7YJC
 
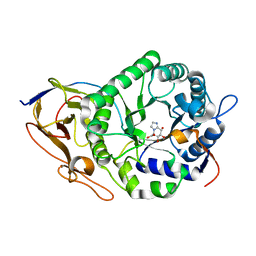 | | Crystal structure of Human HPSE1 in complex with inhibitor | | Descriptor: | (5~{S},6~{R},7~{S},8~{S})-6,7,8-tris(oxidanyl)-5,6,7,8-tetrahydroimidazo[1,2-a]pyridine-5-carboxylic acid, Heparanase 50 kDa subunit, Heparanase 8 kDa subunit | | Authors: | Mima, M, Fujimoto, N, Imai, Y. | | Deposit date: | 2022-07-19 | | Release date: | 2022-12-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Lead identification of novel tetrahydroimidazo[1,2-a]pyridine-5-carboxylic acid derivative as a potent heparanase-1 inhibitor.
Bioorg.Med.Chem.Lett., 79, 2022
|
|
5G2N
 
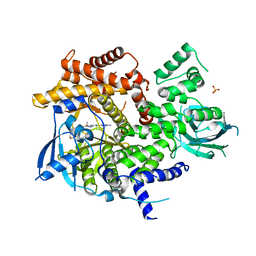 | | X-ray structure of PI3Kinase Gamma in complex with Copanlisib | | Descriptor: | 2-azanyl-~{N}-[7-methoxy-8-(3-morpholin-4-ylpropoxy)-2,3-dihydroimidazo[1,2-c]quinazolin-5-yl]pyrimidine-5-carboxamide, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM, SULFATE ION | | Authors: | Schaefer, M, Scott, W.J, Hentemann, M.F, Rowley, R.B, Bull, C.O, Jenkins, S, Bullion, A.M, Johnson, J, Redman, A, Robbins, A.H, Esler, W, Fracasso, R.P, Garrison, T, Hamilton, M, Michels, M, Wood, J.E, Wilkie, D.P, Xiao, H, Levy, J, Liu, N, Stasik, E, Brands, M, Lefranc, J. | | Deposit date: | 2016-04-11 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Discovery and Sar of Novel 2,3-Dihydroimidazo(1,2-C)Quinazoline Pi3K Inhibitors: Identification of Copanlisib (Bay 80-6946)
Chemmedchem, 11, 2016
|
|
8QSO
 
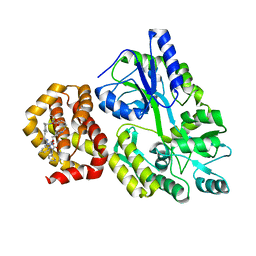 | | Crystal structure of human Mcl-1 in complex with compound 1 | | Descriptor: | (13S,16R,19S)-16-benzyl-43-ethoxy-N-methyl-7,11,14,17-tetraoxo-13-phenyl-5-oxa-2,8,12,15,18-pentaaza-1(1,4),4(1,2)-dibenzena-9(1,4)-cyclohexanacycloicosaphane-19-carboxamide, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Hekking, K.F.W, Gremmen, S, Maroto, S, Keefe, A.D, Zhang, Y. | | Deposit date: | 2023-10-10 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.106 Å) | | Cite: | Development of Potent Mcl-1 Inhibitors: Structural Investigations on Macrocycles Originating from a DNA-Encoded Chemical Library Screen.
J.Med.Chem., 67, 2024
|
|
3UFN
 
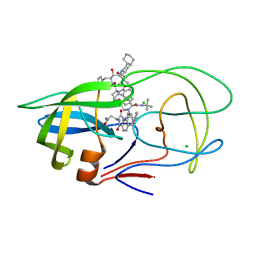 | | Crystal Structure of Multidrug Resistant HIV-1 Protease Clinical Isolate PR20 in Complex with Saquinavir | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, CHLORIDE ION, HIV-1 protease | | Authors: | Agniswamy, J, Chen-Hsiang, S, Aniana, A, Sayer, J.M, Louis, J.M, Weber, I.T. | | Deposit date: | 2011-11-01 | | Release date: | 2012-03-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | HIV-1 protease with 20 mutations exhibits extreme resistance to clinical inhibitors through coordinated structural rearrangements.
Biochemistry, 51, 2012
|
|
4FMZ
 
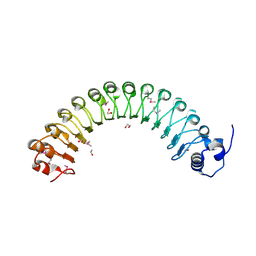 | |
3UH4
 
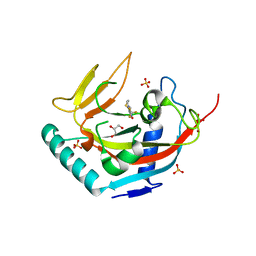 | | TANKYRASE-1 complexed with NVP-XAV939 | | Descriptor: | 2-[4-(trifluoromethyl)phenyl]-7,8-dihydro-5H-thiopyrano[4,3-d]pyrimidin-4-ol, GLYCEROL, SULFATE ION, ... | | Authors: | Stams, T, Kirby, C. | | Deposit date: | 2011-11-03 | | Release date: | 2012-02-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of human tankyrase 1 in complex with small-molecule inhibitors PJ34 and XAV939.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3F53
 
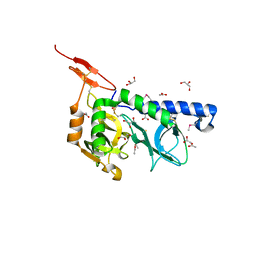 | |
7WPN
 
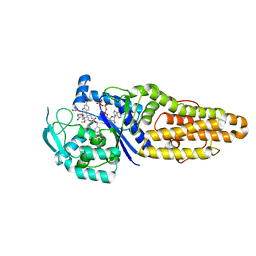 | | Methionyl-tRNA synthetase from Staphylococcus aureus complexed with a phenylbenzimidazole inhibitor and ATP | | Descriptor: | (phenylmethyl) N-[(2R)-2-[2-(4-bromanyl-3-oxidanyl-phenyl)-5-(methylcarbamoyl)benzimidazol-1-yl]-2-(3,4-dimethoxyphenyl)ethyl]carbamate, 1,2-ETHANEDIOL, ACETIC ACID, ... | | Authors: | Yi, J, Cai, Z, Qiu, H, Lu, F, Chen, B, Luo, Z, Gu, Q, Xu, J, Zhou, H. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment screening and structural analyses highlight the ATP-assisted ligand binding for inhibitor discovery against type 1 methionyl-tRNA synthetase.
Nucleic Acids Res., 50, 2022
|
|
3VBV
 
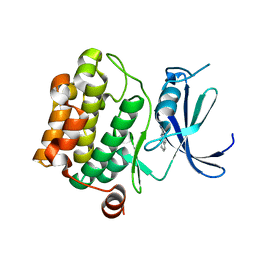 | |
6U6F
 
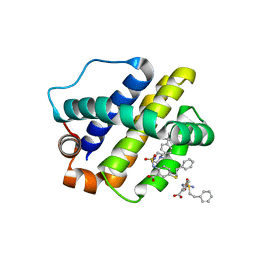 | | The crystal structure of anti-apoptotic Mcl-1 protein in complex with 2, 5-substituted benzoic acid inhibitor 21 | | Descriptor: | 2-[({4-[(4-tert-butylphenyl)methyl]piperazin-1-yl}sulfonyl)amino]-5-[(2-phenylethyl)sulfanyl]benzoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Yang, Y, Stuckey, J.A, Nikolovska-Coleska, Z. | | Deposit date: | 2019-08-29 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery and Characterization of 2,5-Substituted Benzoic Acid Dual Inhibitors of the Anti-apoptotic Mcl-1 and Bfl-1 Proteins.
J.Med.Chem., 63, 2020
|
|
5T2Z
 
 | | Crystal Structure of Multi-drug Resistant HIV-1 Protease PR-S17 in Complex with Darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | Authors: | Agniswamy, J, Weber, I.T. | | Deposit date: | 2016-08-24 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Studies of a Rationally Selected Multi-Drug Resistant HIV-1 Protease Reveal Synergistic Effect of Distal Mutations on Flap Dynamics.
PLoS ONE, 11, 2016
|
|
5E1B
 
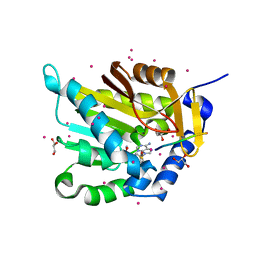 | | Crystal structure of NRMT1 in complex with SPKRIA peptide | | Descriptor: | GLYCEROL, N-terminal Xaa-Pro-Lys N-methyltransferase 1, RCC1, ... | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-29 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for substrate recognition by the human N-terminal methyltransferase 1.
Genes Dev., 29, 2015
|
|
3TC8
 
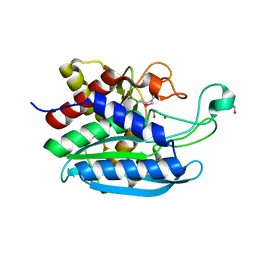 | |
3DS0
 
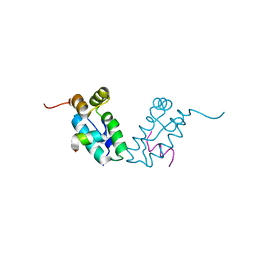 | |
3D61
 
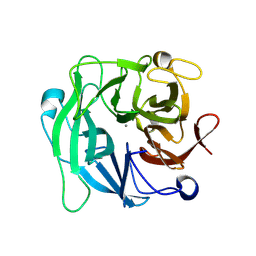 | | Crystal Structure Analysis of 1,5-alpha-arabinanase catalytic mutant (AbnBD147A) complexed to arabinobiose | | Descriptor: | CALCIUM ION, Intracellular arabinanase, alpha-L-arabinofuranose-(1-5)-beta-L-arabinofuranose | | Authors: | Alhassid, A, Ben David, A, Shoham, Y, Shoham, G. | | Deposit date: | 2008-05-18 | | Release date: | 2009-04-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of an inverting GH 43 1,5-alpha-L-arabinanase from Geobacillus stearothermophilus complexed with its substrate
Biochem.J., 422, 2009
|
|
3UZA
 
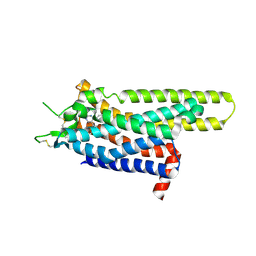 | | Thermostabilised Adenosine A2A receptor in complex with 6-(2,6-Dimethylpyridin-4-yl)-5-phenyl-1,2,4-triazin-3-amine | | Descriptor: | 6-(2,6-dimethylpyridin-4-yl)-5-phenyl-1,2,4-triazin-3-amine, Adenosine receptor A2a | | Authors: | Congreve, M, Andrews, S.P, Dore, A.S, Hollenstein, K, Hurrell, E, Langmead, C.J, Mason, J.S, Ng, I.W, Tehan, B, Zhukov, A, Weir, M, Marshall, F.H. | | Deposit date: | 2011-12-07 | | Release date: | 2012-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.273 Å) | | Cite: | Discovery of 1,2,4-Triazine Derivatives as Adenosine A(2A) Antagonists using Structure Based Drug Design
J.Med.Chem., 55, 2012
|
|
3V5T
 
 | |
3V4T
 
 | | E. cloacae C115D MURA liganded with UNAG | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, UDP-N-acetylglucosamine 1-carboxyvinyltransferase, ... | | Authors: | Zhu, J.-Y, Yang, Y, Schonbrunn, E. | | Deposit date: | 2011-12-15 | | Release date: | 2012-03-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Functional Consequence of Covalent Reaction of Phosphoenolpyruvate with UDP-N-acetylglucosamine 1-Carboxyvinyltransferase (MurA).
J.Biol.Chem., 287, 2012
|
|
3D5Z
 
 | | Crystal Structure Analysis of 1,5-alpha-arabinanase catalytic mutant (AbnBE201A) complexed to arabinotriose | | Descriptor: | CALCIUM ION, Intracellular arabinanase, alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose-(1-5)-beta-L-arabinofuranose | | Authors: | Alhassid, A, Ben David, A, Shoham, Y, Shoham, G. | | Deposit date: | 2008-05-18 | | Release date: | 2009-04-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an inverting GH 43 1,5-alpha-L-arabinanase from Geobacillus stearothermophilus complexed with its substrate
Biochem.J., 422, 2009
|
|
6F20
 
 | | Complex between MTH1 and compound 1 (a 7-azaindole-4-ester derivative) | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, ACETATE ION, Ethyl 1H-pyrrolo[2,3-b]pyridine-4-carboxylate, ... | | Authors: | Viklund, J, Talagas, A, Tresaugues, L, Andersson, M, Ericsson, U, Forsblom, R, Ginman, T, Hallberg, K, Lindstrom, J, Persson, L, Silvander, C, Rahm, F. | | Deposit date: | 2017-11-23 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Creation of a Novel Class of Potent and Selective MutT Homologue 1 (MTH1) Inhibitors Using Fragment-Based Screening and Structure-Based Drug Design.
J. Med. Chem., 61, 2018
|
|
6ECN
 
 | | HIV-1 CA 1/2-hexamer-EE | | Descriptor: | HIV-1 CA | | Authors: | Summers, B.J, Xiong, Y. | | Deposit date: | 2018-08-08 | | Release date: | 2019-08-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Modular HIV-1 Capsid Assemblies Reveal Diverse Host-Capsid Recognition Mechanisms.
Cell Host Microbe, 26, 2019
|
|
7NLD
 
 | | Structure of human Programmed cell death 1 ligand 1 (PD-L1) with low molecular mass inhibitor | | Descriptor: | N-(2-((2'-chloro-3'-(2,3-dihydrobenzo[b][1,4]dioxin-6-yl)-3-methoxy-[1,1'-biphenyl]-4-yl)(methyl)amino)ethyl)methanesulfonamide, Programmed cell death 1 ligand 1 | | Authors: | Sala, D, Magiera-Mularz, K, Muszak, D, Surmiak, E, Grudnik, P, Holak, T.A. | | Deposit date: | 2021-02-22 | | Release date: | 2021-08-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Terphenyl-Based Small-Molecule Inhibitors of Programmed Cell Death-1/Programmed Death-Ligand 1 Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
5AAN
 
 | | Crystal structure of Drosophila NCS-1 bound to penothiazine FD44 | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, CALCIUM ION, CG5907-PA, ... | | Authors: | Chaves-Sanjuan, A, Infantes, L, Sanchez-Barrena, M.J. | | Deposit date: | 2015-07-27 | | Release date: | 2017-01-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Interference of the complex between NCS-1 and Ric8a with phenothiazines regulates synaptic function and is an approach for fragile X syndrome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5FG6
 
 | | Crystal structure of the bromodomain of human BRD1 (BRPF2) in complex with OF-1 chemical probe | | Descriptor: | 1,2-ETHANEDIOL, 4-bromanyl-~{N}-(6-methoxy-1,3-dimethyl-2-oxidanylidene-benzimidazol-5-yl)-2-methyl-benzenesulfonamide, Bromodomain-containing protein 1, ... | | Authors: | Tallant, C, Owen, D.R, Gerstenberger, B.S, Savitsky, P, Chaikuad, A, Fedorov, O, Nunez-Alonso, G, Filippakopoulos, P, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Muller, S, Knapp, S. | | Deposit date: | 2015-12-20 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structure of the bromodomain of human BRD1 (BRPF2) in complex with OF-1 chemical probe
To Be Published
|
|
