5YBJ
 
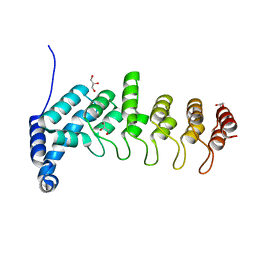 | | Structure of apo KANK1 ankyrin domain | | Descriptor: | GLYCEROL, KN motif and ankyrin repeat domain-containing protein 1 | | Authors: | Guo, Q, Liao, S, Min, J, Xu, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-09-05 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.341 Å) | | Cite: | Structural basis for the recognition of kinesin family member 21A (KIF21A) by the ankyrin domains of KANK1 and KANK2 proteins.
J. Biol. Chem., 293, 2018
|
|
1A3Q
 
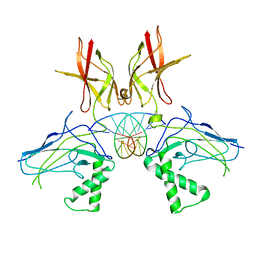 | | HUMAN NF-KAPPA-B P52 BOUND TO DNA | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*AP*AP*TP*CP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*GP*AP*TP*TP*CP*CP*CP*C)-3'), PROTEIN (NUCLEAR FACTOR KAPPA-B P52) | | Authors: | Cramer, P, Larson, C.J, Verdine, G.L, Muller, C.W. | | Deposit date: | 1998-01-23 | | Release date: | 1998-06-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the human NF-kappaB p52 homodimer-DNA complex at 2.1 A resolution.
EMBO J., 16, 1997
|
|
9H8R
 
 | | Crystal structure of Nkp46 in complex with a bicyclic peptide BCY00016132 | | Descriptor: | 1,2-ETHANEDIOL, 1-[3,5-bis(3-bromanylpropanoyl)-1,3,5-triazinan-1-yl]-3-bromanyl-propan-1-one, 2-AMINO-ETHANETHIOL, ... | | Authors: | Pellegrino, S, Carr, K, Bezerra, G.A. | | Deposit date: | 2024-10-29 | | Release date: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of Nkp46 in complex with a bicyclic peptide BCY00016132
To Be Published
|
|
5MKE
 
 | | cryoEM Structure of Polycystin-2 in complex with cations and lipids | | Descriptor: | 1,2-DIPALMITOYL-SN-GLYCERO-3-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wilkes, M, Madej, M.G, Ziegler, C. | | Deposit date: | 2016-12-04 | | Release date: | 2017-01-18 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Molecular insights into lipid-assisted Ca(2+) regulation of the TRP channel Polycystin-2.
Nat. Struct. Mol. Biol., 24, 2017
|
|
6TTN
 
 | | N-terminally truncated hyoscyamine 6-hydroxylase (tH6H) in complex with N-oxalylglycine and hyoscyamine | | Descriptor: | 1,2-ETHANEDIOL, Hyoscyamine 6 beta-hydroxylase, N-OXALYLGLYCINE, ... | | Authors: | Kluza, A, Mrugala, B, Porebski, P.J, Kurpiewska, K, Niedzialkowska, E, Weiss, M.S, Borowski, T. | | Deposit date: | 2019-12-29 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Regioselectivity of hyoscyamine 6 beta-hydroxylase-catalysed hydroxylation as revealed by high-resolution structural information and QM/MM calculations.
Dalton Trans, 49, 2020
|
|
7W0G
 
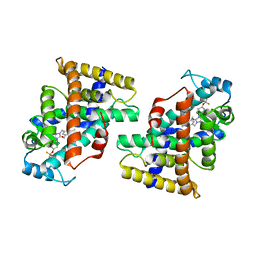 | | Human PPAR delta ligand binding domain in complex with a synthetic agonist H11 | | Descriptor: | 2-[2,6-dimethyl-4-[[5-oxidanylidene-4-[4-(trifluoromethyloxy)phenyl]-1,2,4-triazol-1-yl]methyl]phenoxy]-2-methyl-propanoic acid, Peroxisome proliferator-activated receptor delta | | Authors: | Dai, L, Sun, H.B, Yuan, H.L, Feng, Z.Q. | | Deposit date: | 2021-11-18 | | Release date: | 2022-02-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.443 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Triazolone Derivatives as Potent PPAR alpha / delta Dual Agonists for the Treatment of Nonalcoholic Steatohepatitis.
J.Med.Chem., 65, 2022
|
|
7VB6
 
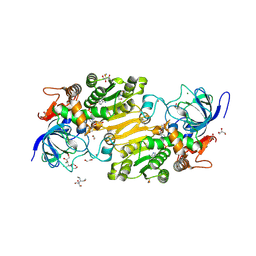 | | Crystal structure of hydroxynitrile lyase from Linum usitatissium complexed with (R)-2-hydroxy-2-methylbutanenitrile | | Descriptor: | (2R)-2-methyl-2-oxidanyl-butanenitrile, 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Zheng, D, Nakabayashi, M, Asano, Y. | | Deposit date: | 2021-08-30 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural characterization of Linum usitatissimum hydroxynitrile lyase: A new cyanohydrin decomposition mechanism involving a cyano-zinc complex.
J.Biol.Chem., 298, 2022
|
|
7Q49
 
 | | Local refinement structure of the N-domain of full-length, monomeric, soluble somatic angiotensin I-converting enzyme | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ZINC ION, ... | | Authors: | Lubbe, L, Sewell, B.T, Sturrock, E.D. | | Deposit date: | 2021-10-29 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Cryo-EM reveals mechanisms of angiotensin I-converting enzyme allostery and dimerization.
Embo J., 41, 2022
|
|
6F3I
 
 | | IRAK4 IN COMPLEX WITH inhibitor | | Descriptor: | (3~{R})-3-[4-[[4-(4-ethanoylpiperazin-1-yl)cyclohexyl]amino]pyrrolo[2,1-f][1,2,4]triazin-5-yl]butanamide, Interleukin-1 receptor-associated kinase 4, SULFATE ION | | Authors: | Xue, Y, Degorce, S.L, Robb, G.R, Ferguson, A.D. | | Deposit date: | 2017-11-28 | | Release date: | 2018-05-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Optimization of permeability in a series of pyrrolotriazine inhibitors of IRAK4.
Bioorg. Med. Chem., 26, 2018
|
|
4QVY
 
 | | yCP beta5-A49T-mutant in complex with bortezomib | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-16 | | Release date: | 2015-02-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
4QWR
 
 | | yCP beta5-C52F mutant in complex with carfilzomib | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-17 | | Release date: | 2015-02-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
7ZAB
 
 | | Sam68 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Nadal, M, Puestes-Prior, P. | | Deposit date: | 2022-03-22 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structure and function analysis of Sam68 and hnRNP A1 synergy in the exclusion of exon 7 from SMN2 transcripts.
Protein Sci., 32, 2023
|
|
1L45
 
 | |
5T6D
 
 | | 2.10 A resolution structure of Norovirus 3CL protease in complex with the dipeptidyl inhibitor 7l (hexagonal form) | | Descriptor: | 3-cyclohexyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-{[3-(4-methoxyphenoxy)propyl]sulfonyl}-L- alaninamide, Genome polyprotein | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Kim, Y, Rathnayake, A.D, Damalanka, V.C, Weerawarna, P.M, Doyle, S.T, Alsoudi, A.F, Dissanayake, D.M.P, Chang, K.-O, Groutas, W.C. | | Deposit date: | 2016-09-01 | | Release date: | 2016-11-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based exploration and exploitation of the S4 subsite of norovirus 3CL protease in the design of potent and permeable inhibitors.
Eur J Med Chem, 126, 2016
|
|
6AOK
 
 | | Crystal structure of Legionella pneumophila effector Ceg4 with N-terminal TEV protease cleavage sequence | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Ceg4, ... | | Authors: | Stogios, P.J, Cuff, M.E, Nocek, B, Evdokimova, E, Egorova, O, Yim, V, Di Leo, R, Savchenko, A. | | Deposit date: | 2017-08-16 | | Release date: | 2018-01-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | TheLegionella pneumophilaeffector Ceg4 is a phosphotyrosine phosphatase that attenuates activation of eukaryotic MAPK pathways.
J. Biol. Chem., 293, 2018
|
|
1L55
 
 | |
6EZJ
 
 | | Imidazoleglycerol-phosphate dehydratase | | Descriptor: | Imidazoleglycerol-phosphate dehydratase 2, chloroplastic, MANGANESE (II) ION, ... | | Authors: | Rawson, S, Bisson, C, Hurdiss, D.L, Muench, S.P. | | Deposit date: | 2017-11-15 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Elucidating the structural basis for differing enzyme inhibitor potency by cryo-EM.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6TTK
 
 | | Crystal structure of the kelch domain of human KLHL12 in complex with DVL1 peptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DVL1, ... | | Authors: | Chen, Z, Williams, E, Pike, A.C.W, Strain-Damerell, C, Wang, D, Chalk, R, Burgess-Brown, N, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2019-12-27 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.383 Å) | | Cite: | Identification of a PGXPP degron motif in dishevelled and structural basis for its binding to the E3 ligase KLHL12.
Open Biology, 10, 2020
|
|
7Q1M
 
 | | Crystal structure of human butyrylcholinesterase in complex with N-[(2S)-3-[(cyclohexylmethyl)amino]-2-hydroxypropyl]-2,2-diphenylacetamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Brazzolotto, X, Panek, D, Pasieka, A, Malawska, B, Nachon, F. | | Deposit date: | 2021-10-20 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Discovery of new, highly potent and selective inhibitors of BuChE - design, synthesis, in vitro and in vivo evaluation and crystallography studies.
Eur.J.Med.Chem., 249, 2023
|
|
6AS5
 
 | | CRYSTAL STRUCTURE OF PROTEIN CitE FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH MAGNESIUM, ACETOACETATE AND COENZYME A | | Descriptor: | ACETOACETIC ACID, COENZYME A, Citrate lyase subunit beta-like protein, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Wang, H, Bonanno, J.B, Carvalho, L, Almo, S.C. | | Deposit date: | 2017-08-23 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.035 Å) | | Cite: | An essential bifunctional enzyme inMycobacterium tuberculosisfor itaconate dissimilation and leucine catabolism.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
9KFJ
 
 | | Cryo-EM structure of the compound 4-bound human relaxin family peptide receptor 3 (RXFP3)-Gi complex | | Descriptor: | 1-[2-(4-chlorophenyl)ethyl]-3-[(7-ethyl-5-oxidanyl-1H-indol-3-yl)methylideneamino]guanidine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Chen, Y, Zhou, Q.T, Yan, S.Y, Yan, J.H, Yang, D.H, Chen, J, Wang, M.W. | | Deposit date: | 2024-11-06 | | Release date: | 2025-08-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular mechanism underlying non-discriminatory recognition of relaxin-3 by RXFP3 and RXFP4.
Commun Biol, 8, 2025
|
|
9HGI
 
 | | HSV-1 Origin Binding Protein in complex with double-stranded DNA recognition sequence OriS with 6 basepairs removed from the AT-rich region | | Descriptor: | DNA (5'-D(P*AP*AP*GP*CP*GP*TP*TP*CP*GP*CP*AP*CP*TP*TP*CP*GP*TP*CP*CP*CP*AP*AP*TP*A)-3'), DNA (5'-D(P*TP*AP*TP*TP*GP*GP*GP*AP*CP*GP*AP*AP*GP*TP*GP*CP*GP*AP*AP*CP*GP*CP*TP*T)-3'), Replication origin-binding protein | | Authors: | Gustavsson, E, Grunewald, K, Elias, P, Hallberg, B.M. | | Deposit date: | 2024-11-19 | | Release date: | 2025-10-22 | | Last modified: | 2025-11-05 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | The herpes simplex origin-binding protein: mechanisms for sequence-specific DNA binding and dimerization revealed by Cryo-EM.
Nucleic Acids Res., 53, 2025
|
|
9HM2
 
 | | A swapped dimeric form of ZO1/TJP1 PDZ2 in complex with the C-terminal peptide from protein E of SARS-CoV-2 | | Descriptor: | Envelope small membrane protein, IODIDE ION, Tight junction protein ZO-1 | | Authors: | Alvarez, F, Mechaly, A, Haouz, A, Caillet-Saguy, C. | | Deposit date: | 2024-12-06 | | Release date: | 2025-10-22 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | HTRF-based identification of small molecules targeting SARS-CoV-2 E protein interaction with ZO-1 PDZ2
Biorxiv, 2025
|
|
7Q1N
 
 | | Crystal structure of human butyrylcholinesterase in complex with N-[(2R)-3-[(cyclohexylmethyl)amino]-2-hydroxypropyl]-2,2-diphenylacetamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Brazzolotto, X, Panek, D, Pasieka, A, Malawska, B, Nachon, F. | | Deposit date: | 2021-10-20 | | Release date: | 2022-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of new, highly potent and selective inhibitors of BuChE - design, synthesis, in vitro and in vivo evaluation and crystallography studies.
Eur.J.Med.Chem., 249, 2023
|
|
7VEK
 
 | | Crystal structure of Phytolacca americana UGT3 with capsaicin and UDP-2fluoroglucose | | Descriptor: | (6E)-N-(4-hydroxy-3-methoxybenzyl)-8-methylnon-6-enamide, 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, Glycosyltransferase, ... | | Authors: | Maharjan, R, Fukuda, Y, Nakayama, T, Nakayama, T, Hamada, H, Ozaki, S, Inoue, T. | | Deposit date: | 2021-09-09 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for substrate recognition in the Phytolacca americana glycosyltransferase PaGT3.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
