6VL8
 
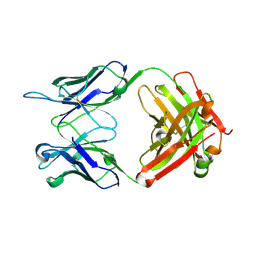 | | Anti-PEG antibody 6-3 Fab fragment in complex with PEG | | Descriptor: | 1,2-ETHANEDIOL, 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, 6-3 Fab heavy chain, ... | | Authors: | Nicely, N.I, Huckaby, J.T, Lai, S.K, Jacobs, T.M. | | Deposit date: | 2020-01-23 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structure of an anti-PEG antibody reveals an open ring that captures highly flexible PEG polymers
Commun Chem, 3, 2020
|
|
8QH9
 
 | |
6PPA
 
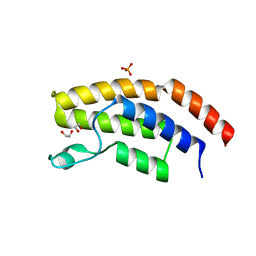 | | Crystal structure of the unliganded bromodomain of human BRD7 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 7, PHOSPHATE ION | | Authors: | Chan, A, Karim, M.R, Zhu, J, Schonbrunn, E. | | Deposit date: | 2019-07-05 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
8JK1
 
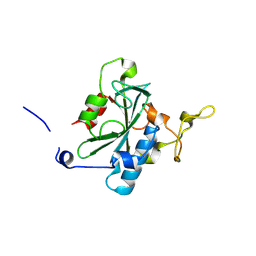 | | Crystal structure of QA-hNTAQ1 C28S | | Descriptor: | Protein N-terminal glutamine amidohydrolase | | Authors: | Kang, J.M, Han, B.W. | | Deposit date: | 2023-05-31 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.067 Å) | | Cite: | Structural study for substrate recognition of human N-terminal glutamine amidohydrolase 1 in the arginine N-degron pathway.
Protein Sci., 33, 2024
|
|
8C70
 
 | | Pyrrolidine fragment 1 bound to endothiapepsin | | Descriptor: | (3~{R},4~{R})-4-[4-[(5-bromanylpyridin-3-yl)oxymethyl]-1,2,3-triazol-1-yl]pyrrolidin-3-ol, Endothiapepsin | | Authors: | Wiese, J.N, Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2023-01-12 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Fragtory: Pharmacophore-Focused Design, Synthesis, and Evaluation of an sp 3 -Enriched Fragment Library.
J.Med.Chem., 66, 2023
|
|
8C6T
 
 | | Fragment screening hit IV bound to endothiapepsin | | Descriptor: | 1-[3,5-bis(chloranyl)phenoxy]propan-2-amine, Endothiapepsin, GLYCEROL | | Authors: | Wiese, J.N, Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2023-01-12 | | Release date: | 2023-05-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Fragtory: Pharmacophore-Focused Design, Synthesis, and Evaluation of an sp 3 -Enriched Fragment Library.
J.Med.Chem., 66, 2023
|
|
6VPM
 
 | |
7OO4
 
 | |
8E6K
 
 | | 2H08 Fab in complex with influenza virus neuraminidase from A/Brevig Mission/1/1918 (H1N1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2H08 fragment antigen binding heavy chain, 2H08 fragment antigen binding light chain, ... | | Authors: | Turner, H.L, Ozorowski, G, Ward, A.B. | | Deposit date: | 2022-08-22 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Human anti-N1 monoclonal antibodies elicited by pandemic H1N1 virus infection broadly inhibit HxN1 viruses in vitro and in vivo.
Immunity, 56, 2023
|
|
8DZZ
 
 | | Cryo-EM structure of chi dynein bound to Lis1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Dynein heavy chain, ... | | Authors: | Reimer, J.M, Lahiri, I, Leschziner, A.E. | | Deposit date: | 2022-08-08 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Lis1 relieves cytoplasmic dynein-1 autoinhibition by acting as a molecular wedge.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7QUB
 
 | | EV-A71-3Cpro in complex with inhibitor MG78 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, Protease 3C, SODIUM ION | | Authors: | El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2022-01-17 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | From Repurposing to Redesign: Optimization of Boceprevir to Highly Potent Inhibitors of the SARS-CoV-2 Main Protease.
Molecules, 27, 2022
|
|
9FRM
 
 | | The crystal structure of glycogen phosphorylase with an indole derivative | | Descriptor: | (7~{S})-1-(4-hydroxyphenyl)-2-[(4-hydroxyphenyl)amino]-6,6-dimethyl-7-oxidanyl-7,8-dihydrobenzo[cd]indole-3-carboxylic acid, DIMETHYL SULFOXIDE, Glycogen phosphorylase, ... | | Authors: | Koulas, S.M, Leonidas, D.D. | | Deposit date: | 2024-06-19 | | Release date: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis and evaluation of tetrahydrobenzo[cd]indole derivatives as glycogen phosphorylase inhibitors
Med.Chem.Res., 2025
|
|
7QL8
 
 | | SARS-COV2 Main Protease in complex with inhibitor MG78 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2021-12-19 | | Release date: | 2022-04-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.807 Å) | | Cite: | From Repurposing to Redesign: Optimization of Boceprevir to Highly Potent Inhibitors of the SARS-CoV-2 Main Protease.
Molecules, 27, 2022
|
|
6Y1B
 
 | | X-ray structure of Lactobacillus brevis alcohol dehydrogenase mutant K32A_Q126K | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Hermann, J, Bischoff, D, Janowski, R, Niessing, D, Grob, P, Hekmat, D, Weuster-Botz, D. | | Deposit date: | 2020-02-11 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Contact Engineering Enables Efficient Capture and Purification of an Oxidoreductase by Technical Crystallization.
Biotechnol J, 15, 2020
|
|
6PGU
 
 | |
7A6Z
 
 | | Structure of P226G BlaC from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, GLYCEROL, ... | | Authors: | Chikunova, A, Ahmad, M.U, Perrakis, A, Ubbink, M. | | Deposit date: | 2020-08-27 | | Release date: | 2021-10-06 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Conserved proline residues prevent dimerization and aggregation in the beta-lactamase BlaC.
Protein Sci., 33, 2024
|
|
8XVR
 
 | | Crystal structure of inulosucrase from Lactobacillus reuteri 121 mutant R544W | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ni, D, Hou, X, Cheng, M, Xu, W, Rao, Y, Mu, W. | | Deposit date: | 2024-01-15 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structure-guided tunnel engineering to reveal the molecular basis of sugar chain extension of inulosucrase
To Be Published
|
|
8VLS
 
 | | Structure of VCP in complex with an ATPase activator (D2 domains only, dodecameric form) | | Descriptor: | (3R)-N-[2-(ethylsulfanyl)phenyl]-3-(1-oxo-1,3-dihydro-2H-isoindol-2-yl)butanamide, Transitional endoplasmic reticulum ATPase | | Authors: | Jones, N.H, Urnivicius, L, Kapoor, T.M. | | Deposit date: | 2024-01-12 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Allosteric activation of VCP, an AAA unfoldase, by small molecule mimicry.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7QGZ
 
 | | Human carbonic anhydrase II in complex with Methyl 3-((4-methylthiazol-2-yl)(4-sulfamoylphenyl)amino)propanoate | | Descriptor: | Carbonic anhydrase 2, ZINC ION, methyl 3-[(4-methyl-1,3-thiazol-2-yl)-(4-sulfamoylphenyl)amino]propanoate | | Authors: | Paketuryte-Latve, V, Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2021-12-10 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Beta and Gamma Amino Acid-Substituted Benzenesulfonamides as Inhibitors of Human Carbonic Anhydrases.
Pharmaceuticals, 15, 2022
|
|
6MNP
 
 | | Crystal structure of KPC-2 with compound 6 | | Descriptor: | 3-(1H-tetrazol-5-ylmethyl)-5,6,7,8-tetrahydro[1]benzothieno[2,3-d]pyrimidin-4(3H)-one, Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL | | Authors: | Akhtar, A, Chen, Y. | | Deposit date: | 2018-10-02 | | Release date: | 2019-04-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Active-Site Druggability of Carbapenemases and Broad-Spectrum Inhibitor Discovery.
Acs Infect Dis., 5, 2019
|
|
7QGY
 
 | | Human carbonic anhydrase II in complex with 3-((5-(ethoxycarbonyl)-4-methylthiazol-2-yl)(4-sulfamoylphenyl)amino)propanoic acid | | Descriptor: | 3-[(5-ethoxycarbonyl-4-methyl-1,3-thiazol-2-yl)-(4-sulfamoylphenyl)amino]propanoic acid, Carbonic anhydrase 2, ZINC ION | | Authors: | Paketuryte-Latve, V, Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2021-12-10 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Beta and Gamma Amino Acid-Substituted Benzenesulfonamides as Inhibitors of Human Carbonic Anhydrases.
Pharmaceuticals, 15, 2022
|
|
6G34
 
 | | Crystal structure of haspin in complex with 5-iodotubercidin | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, IODIDE ION, ... | | Authors: | Heroven, C, Chaikuad, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-24 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Halogen-Aromatic pi Interactions Modulate Inhibitor Residence Times.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6MPS
 
 | | TagT bound to LIIa-WTA | | Descriptor: | 2-(acetylamino)-4-O-[2-(acetylamino)-2-deoxy-beta-D-mannopyranosyl]-2-deoxy-1-O-[(S)-{[(R)-{[(2Z,6Z,10Z,14E,18E)-3,7,11,15,19,23-hexamethyltetracosa-2,6,10,14,18,22-hexaen-1-yl]oxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]-alpha-D-glucopyranose, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Owens, T.W, Schaefer, K, Kahne, D, Walker, S. | | Deposit date: | 2018-10-08 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Substrate Preferences Establish the Order of Cell Wall Assembly in Staphylococcus aureus.
J. Am. Chem. Soc., 140, 2018
|
|
6VK5
 
 | | Crystal Structure of Methylosinus trichosporium OB3b Soluble Methane Monooxygenase Hydroxylase and Regulatory Component Complex | | Descriptor: | 1,2-ETHANEDIOL, BENZOIC ACID, CHLORIDE ION, ... | | Authors: | Jones, J.C, Banerjee, R, Shi, K, Aihara, H, Lipscomb, J.D. | | Deposit date: | 2020-01-18 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural Studies of theMethylosinus trichosporiumOB3b Soluble Methane Monooxygenase Hydroxylase and Regulatory Component Complex Reveal a Transient Substrate Tunnel.
Biochemistry, 59, 2020
|
|
7O1U
 
 | |
