8CFR
 
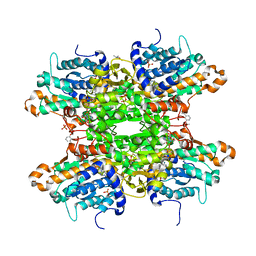 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with F2X-Entry library fragment G03 | | Descriptor: | 3-ethoxybenzene-1-carboximidamide, ADENINE, Adenosylhomocysteinase, ... | | Authors: | Malecki, P.H, Gawel, M, Stepniewska, M, Brzezinski, K. | | Deposit date: | 2023-02-03 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with fragment F2X-Entry G03
To be published
|
|
8CFY
 
 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with F2X-Entry library fragment H08 | | Descriptor: | ADENINE, Adenosylhomocysteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Malecki, P.H, Gawel, M, Stepniewska, M, Brzezinski, K. | | Deposit date: | 2023-02-03 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with fragment F2X-Entry H08
To be published
|
|
8CFB
 
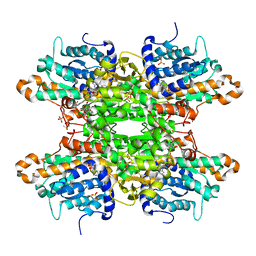 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with F2X-Entry library fragment D02 | | Descriptor: | ADENINE, Adenosylhomocysteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Malecki, P.H, Gawel, M, Stepniewska, M, Brzezinski, K. | | Deposit date: | 2023-02-03 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with F2X-Entry library fragment D02
To be published
|
|
8CFO
 
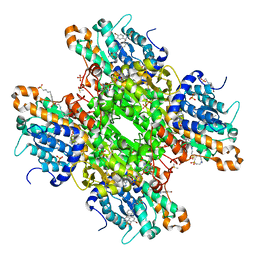 | | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with F2X-Entry library fragment F04 | | Descriptor: | ADENINE, Adenosylhomocysteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Malecki, P.H, Gawel, M, Stepniewska, M, Brzezinski, K. | | Deposit date: | 2023-02-03 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structure of S-adenosyl-L-homocysteine hydrolase from P. aeruginosa in complex with fragment F2X-Entry F04
To be published
|
|
6W1W
 
 | | Crystal Structure of Motility Associated Killing Factor B from Vibrio cholerae | | Descriptor: | 1,2-ETHANEDIOL, motility-associated killing factor MakB | | Authors: | Kim, Y, Welk, L, Jedrzejczak, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-03-04 | | Release date: | 2020-03-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | A Genomic Island of Vibrio cholerae Encodes a Three-Component Cytotoxin with Monomer and Protomer Forms Structurally Similar to Alpha-Pore-Forming Toxins.
J.Bacteriol., 204, 2022
|
|
8CX8
 
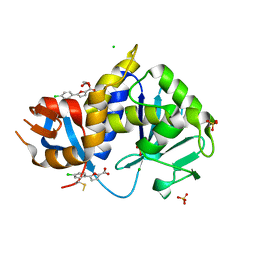 | | Stx2A1-BTB13086 | | Descriptor: | 1,2-ETHANEDIOL, 5-(4-chlorophenyl)furan-2-carboxylic acid, CHLORIDE ION, ... | | Authors: | Rudolph, M.J, Li, X.P. | | Deposit date: | 2022-05-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Fragment Screening to Identify Inhibitors Targeting Ribosome Binding of Shiga Toxin 2.
Acs Infect Dis., 10, 2024
|
|
7P3D
 
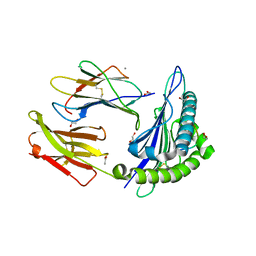 | | MHC I A02 Allele presenting YLQPRTFLL | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Beta-2-microglobulin, ... | | Authors: | Rizkallah, P.J, Sewell, A.K, Wall, A, Fuller, A. | | Deposit date: | 2021-07-07 | | Release date: | 2021-07-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Emergence of immune escape at dominant SARS-CoV-2 killer T cell epitope.
Cell, 185, 2022
|
|
8TGA
 
 | |
8CNA
 
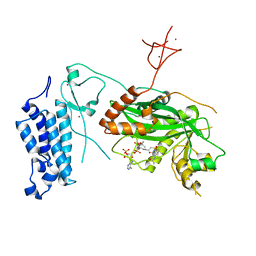 | | Crystal structure of CREBBP-R1446C histone acetyltransferase domain in complex with a bisubstrate inhibitor, Lys-CoA | | Descriptor: | ZINC ION, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]methyl (3R,20R)-20-carbamoyl-3-hydroxy-2,2-dimethyl-4,8,14,22-tetraoxo-12-thia-5,9,15,21-tetraazatricos-1-yl dihydrogen diphosphate, histone acetyltransferase | | Authors: | Mechaly, A.E, Zhang, W, Haouz, A, Green, M, Rodrigues-Lima, F. | | Deposit date: | 2023-02-22 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.463 Å) | | Cite: | Crystal structure of CREBBP-R1446C histone acetyltransferase domain in complex with a bisubstrate inhibitor, Lys-CoA
To Be Published
|
|
6NSK
 
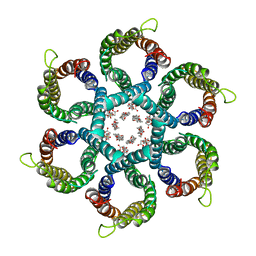 | | CryoEM structure of Helicobacter pylori urea channel in open state. | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, Acid-activated urea channel | | Authors: | Cui, Y.X, Zhou, K, Strugatsky, D, Wen, Y, Sachs, G, Munson, K, Zhou, Z.H. | | Deposit date: | 2019-01-24 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | pH-dependent gating mechanism of theHelicobacter pyloriurea channel revealed by cryo-EM.
Sci Adv, 5, 2019
|
|
8C6N
 
 | |
8C6M
 
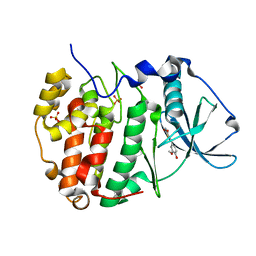 | |
8Z4R
 
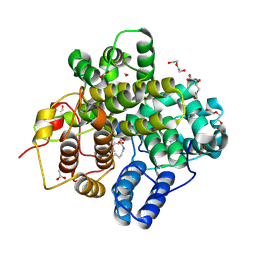 | | The crystal structure of a Hydroquinone Dioxygenase PaD with substrate | | Descriptor: | 2-methoxy-6-methyl-benzene-1,4-diol, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Liu, Z.W, Huang, J.-W, Wang, Y.X, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2024-04-17 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Substrate specificity of a branch of aromatic dioxygenases determined by three distinct motifs.
Nat Commun, 15, 2024
|
|
8C6V
 
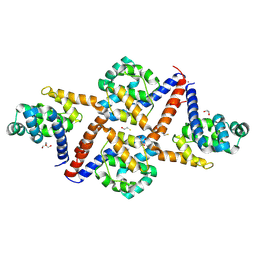 | |
8KI4
 
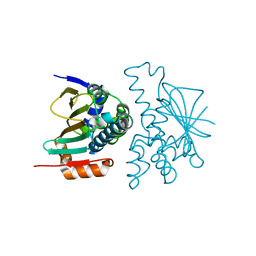 | | Crystal structure of human HSP90 in intermediate state | | Descriptor: | 1,2-ETHANEDIOL, Heat shock protein HSP 90-alpha | | Authors: | Xu, C, Zhang, X.L, Bai, F. | | Deposit date: | 2023-08-22 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Accurate Characterization of Binding Kinetics and Allosteric Mechanisms for the HSP90 Chaperone Inhibitors Using AI-Augmented Integrative Biophysical Studies.
Jacs Au, 4, 2024
|
|
7M4E
 
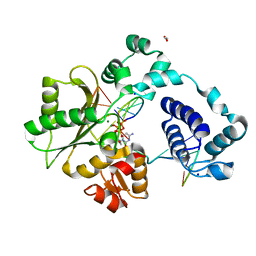 | | DNA Polymerase Lambda, dCTP:At Mg2+ Reaction State Ternary Complex, 120 min | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA (5'-D(*CP*AP*GP*TP*AP*CP*C)-3'), ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2021-03-21 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Watching right and wrong nucleotide insertion captures hidden polymerase fidelity checkpoints.
Nat Commun, 13, 2022
|
|
7QVN
 
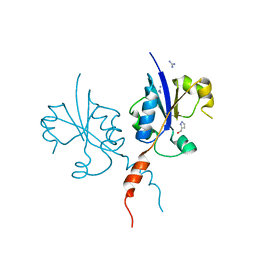 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with(1H-pyrazol-5-yl)methanol | | Descriptor: | 1~{H}-pyrazol-5-ylmethanol, Cholinephosphate cytidylyltransferase, Guanidinium | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2022-01-22 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors
To Be Published
|
|
8WJY
 
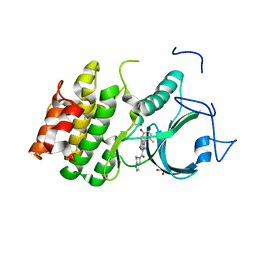 | | PKMYT1_Cocrystal_Cpd 4 | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase, ... | | Authors: | Wang, Y, Wang, C, Liu, T, Qi, H, Chen, S, Cai, X, Zhang, M, Aliper, A, Ren, F, Ding, X, Zhavoronkov, A. | | Deposit date: | 2023-09-26 | | Release date: | 2023-11-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Discovery of Tetrahydropyrazolopyrazine Derivatives as Potent and Selective MYT1 Inhibitors for the Treatment of Cancer.
J.Med.Chem., 67, 2024
|
|
6NIC
 
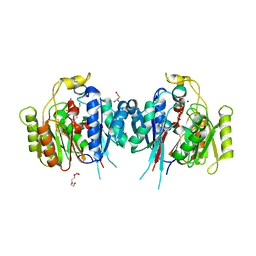 | | Crystal Structure of Medicago truncatula Agmatine Iminohydrolase (Deiminase) in Complex with 6-aminohexanamide | | Descriptor: | 1,2-ETHANEDIOL, 6-aminohexanamide, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sekula, B, Dauter, Z. | | Deposit date: | 2018-12-27 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Study of Agmatine Iminohydrolase FromMedicago truncatula, the Second Enzyme of the Agmatine Route of Putrescine Biosynthesis in Plants.
Front Plant Sci, 10, 2019
|
|
7OZ1
 
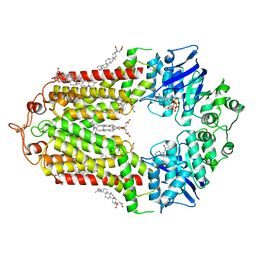 | | Cryo-EM structure of ABCG1 E242Q mutant with ATP and cholesteryl hemisuccinate bound | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-TRIPHOSPHATE, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Skarda, L, Kowal, J, Locher, K.P. | | Deposit date: | 2021-06-25 | | Release date: | 2021-09-22 | | Last modified: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of the Human Cholesterol Transporter ABCG1.
J.Mol.Biol., 433, 2021
|
|
6NYV
 
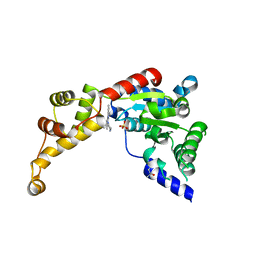 | | Structure of spastin AAA domain in complex with a quinazoline-based inhibitor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, N-(3-tert-butyl-1H-pyrazol-5-yl)-2-[(2R)-2-methylpiperazin-1-yl]quinazolin-4-amine, SULFATE ION, ... | | Authors: | Pisa, R, Cupido, T, Kapoor, T.M. | | Deposit date: | 2019-02-12 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.425 Å) | | Cite: | Designing Allele-Specific Inhibitors of Spastin, a Microtubule-Severing AAA Protein.
J. Am. Chem. Soc., 141, 2019
|
|
8EXN
 
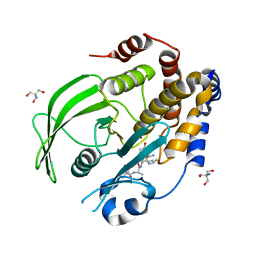 | | Crystal structure of PTP1B D181A/Q262A phosphatase domain with TYK2 activation loop phosphopeptide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Non-receptor tyrosine-protein kinase TYK2 activation loop peptide, PHOSPHATE ION, ... | | Authors: | Morris, R, Kershaw, N.J, Babon, J.J. | | Deposit date: | 2022-10-25 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Structure guided studies of the interaction between PTP1B and JAK.
Commun Biol, 6, 2023
|
|
8EXM
 
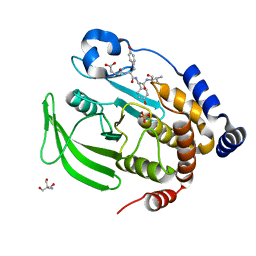 | | Crystal structure of PTP1B D181A/Q262A phosphatase domain with a JAK3 activation loop phosphopeptide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PHOSPHATE ION, Tyrosine-protein kinase JAK3 activation loop peptide, ... | | Authors: | Morris, R, Kershaw, N.J, Babon, J.J. | | Deposit date: | 2022-10-25 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Structure guided studies of the interaction between PTP1B and JAK.
Commun Biol, 6, 2023
|
|
7AJ9
 
 | |
6W11
 
 | |
