6KCF
 
 | |
6HNC
 
 | | Trypanosoma brucei PTR1 in complex with cycloguanil | | Descriptor: | 1-(4-chlorophenyl)-6,6-dimethyl-1,6-dihydro-1,3,5-triazine-2,4-diamine, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Landi, G, Pozzi, C, Mangani, S. | | Deposit date: | 2018-09-14 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Insights into the Development of Cycloguanil Derivatives asTrypanosoma bruceiPteridine-Reductase-1 Inhibitors.
Acs Infect Dis., 5, 2019
|
|
6HOW
 
 | | Trypanosoma brucei PTR1 in complex with the triazine inhibitor 2a (F219). | | Descriptor: | (2~{R})-1-(3,4-dichlorophenyl)-2-(4-nitrophenyl)-2~{H}-1,3,5-triazine-4,6-diamine, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pteridine reductase | | Authors: | Landi, G, Pozzi, C, Mangani, S. | | Deposit date: | 2018-09-18 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Insights into the Development of Cycloguanil Derivatives asTrypanosoma bruceiPteridine-Reductase-1 Inhibitors.
Acs Infect Dis., 5, 2019
|
|
6HQG
 
 | | Cytochrome P450-153 from Phenylobacterium zucineum | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fiorentini, F, Mattevi, A. | | Deposit date: | 2018-09-25 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Extreme Structural Plasticity in the CYP153 Subfamily of P450s Directs Development of Designer Hydroxylases.
Biochemistry, 57, 2018
|
|
6HZA
 
 | |
6IBY
 
 | | Human PFKFB3 in complex with a N-Aryl 6-Aminoquinoxaline inhibitor 6 | | Descriptor: | 3-[[8-(1-methylindol-6-yl)quinoxalin-6-yl]amino]-~{N}-[(3~{S})-1-methylpyrrolidin-3-yl]pyridine-4-carboxamide, 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3, ... | | Authors: | Banaszak, K, Pawlik, H, Bialas, A, Fabritius, C.H, Nowak, M. | | Deposit date: | 2018-12-01 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Synthesis of amide and sulfonamide substituted N-aryl 6-aminoquinoxalines as PFKFB3 inhibitors with improved physicochemical properties.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
7F0H
 
 | |
5UZ4
 
 | | The cryo-EM structure of YjeQ bound to the 30S subunit suggests a fidelity checkpoint function for this protein in ribosome assembly | | Descriptor: | 16S RIBOSOMAL RNA, 3'-O-(N-methylanthraniloyl)-beta:gamma-imidoguanosine-5'-triphosphate, 30S ribosomal protein S10, ... | | Authors: | Razi, A, Guarne, A, Ortega, J. | | Deposit date: | 2017-02-24 | | Release date: | 2017-04-19 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | The cryo-EM structure of YjeQ bound to the 30S subunit suggests a fidelity checkpoint function for this protein in ribosome assembly.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4J6X
 
 | |
6KK7
 
 | | Structure of thermal-stabilised(M6) human GLP-1 receptor transmembrane domain | | Descriptor: | Glucagon-like peptide 1 receptor,Endolysin,Glucagon-like peptide 1 receptor, N-{4-[(R)-(3,3-dimethylcyclobutyl)({6-[4-(trifluoromethyl)-1H-imidazol-1-yl]pyridin-3-yl}amino)methyl]benzene-1-carbonyl}-beta-alanine | | Authors: | Song, G. | | Deposit date: | 2019-07-23 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Mutagenesis facilitated crystallization of GLP-1R.
Iucrj, 6, 2019
|
|
6KK1
 
 | | Structure of thermal-stabilised(M8) human GLP-1 receptor transmembrane domain | | Descriptor: | Glucagon-like peptide 1 receptor,Endolysin,Glucagon-like peptide 1 receptor, N-{4-[(R)-(3,3-dimethylcyclobutyl)({6-[4-(trifluoromethyl)-1H-imidazol-1-yl]pyridin-3-yl}amino)methyl]benzene-1-carbonyl}-beta-alanine | | Authors: | Song, G. | | Deposit date: | 2019-07-23 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mutagenesis facilitated crystallization of GLP-1R.
Iucrj, 6, 2019
|
|
2NY8
 
 | |
7JGI
 
 | | NMR structure of the cNTnC-cTnI chimera bound to A7 | | Descriptor: | 7-{[(5-chloronaphthalen-1-yl)sulfonyl]amino}heptanoic acid, CALCIUM ION, Troponin C, ... | | Authors: | Cai, F, Robertson, I.M, Kampourakis, T, Klein, B.A, Sykes, B.D. | | Deposit date: | 2020-07-19 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Role of Electrostatics in the Mechanism of Cardiac Thin Filament Based Sensitizers.
Acs Chem.Biol., 15, 2020
|
|
4J6Y
 
 | |
4J6W
 
 | | Crystal structure of HFQ from Pseudomonas aeruginosa in complex with CTP | | Descriptor: | CHLORIDE ION, CYTIDINE-5'-DIPHOSPHATE, CYTIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Nikulin, A.D, Murina, V, Lekontseva, N. | | Deposit date: | 2013-02-12 | | Release date: | 2013-07-31 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hfq binds ribonucleotides in three different RNA-binding sites.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2R2L
 
 | | Structure of Farnesyl Protein Transferase bound to PB-93 | | Descriptor: | FARNESYL DIPHOSPHATE, Farnesyltransferase subunit alpha, Farnesyltransferase subunit beta, ... | | Authors: | Strickland, C.O, Voorhis, W. | | Deposit date: | 2007-08-27 | | Release date: | 2008-03-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Efficacy, pharmacokinetics, and metabolism of tetrahydroquinoline inhibitors of Plasmodium falciparum protein farnesyltransferase.
Antimicrob.Agents Chemother., 51, 2007
|
|
4J5Y
 
 | | Crystal structure of Hfq from Pseudomonas aeruginosa in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, Protein hfq, ... | | Authors: | Murina, V, Lekontseva, N, Nikulin, A. | | Deposit date: | 2013-02-11 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.0953 Å) | | Cite: | Hfq binds ribonucleotides in three different RNA-binding sites.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
7UL1
 
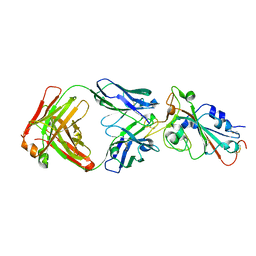 | | Crystal structure of SARS-CoV-2 RBD in complex with the neutralizing IGHV3-53-encoded antibody EH3 isolated from a nonvaccinated pediatric patient | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of EH3, Light chain of EH3, ... | | Authors: | Chen, Y, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2022-04-03 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Molecular basis for antiviral activity of two pediatric neutralizing antibodies targeting SARS-CoV-2 Spike RBD.
Iscience, 26, 2023
|
|
7UL0
 
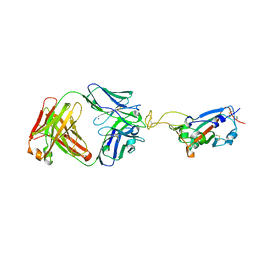 | | Crystal structure of SARS-CoV-2 RBD in complex with the ridge-binding nAb EH8 isolated from a nonvaccinated pediatric patient | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of EH8, ... | | Authors: | Chen, Y, Tolbert, W.D, Bai, X, Pazgier, M. | | Deposit date: | 2022-04-03 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Molecular basis for antiviral activity of two pediatric neutralizing antibodies targeting SARS-CoV-2 Spike RBD.
Iscience, 26, 2023
|
|
7UAH
 
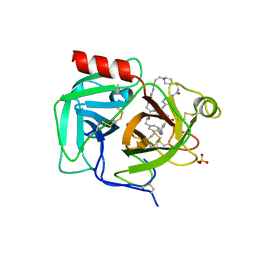 | | Macrocyclic plasmin inhibitor | | Descriptor: | (2~{R})-butane-1,2-diol, (6S,9R,19S,22R)-N-{[4-(aminomethyl)phenyl]methyl}-22-[(3-chlorobenzene-1-sulfonyl)amino]-3,12,21-trioxo-2,6,9,13,20-pentaazatetracyclo[22.2.2.2~6,9~.2~14,17~]dotriaconta-1(26),14,16,24,27,29-hexaene-19-carboxamide, Plasminogen, ... | | Authors: | Guojie, W. | | Deposit date: | 2022-03-12 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Synthesis and Structural Characterization of Macrocyclic Plasmin Inhibitors.
Chemmedchem, 18, 2023
|
|
7UKL
 
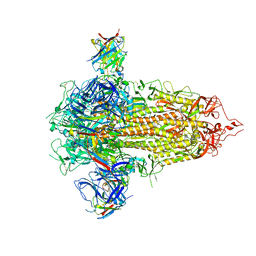 | |
7UQC
 
 | |
7UHC
 
 | |
7UHB
 
 | |
7VKM
 
 | | Crystal structure of TrkA (G595R) kinase domain | | Descriptor: | Tyrosine-protein kinase receptor | | Authors: | Murray, B.W, Rogers, E, Zhai, D, Deng, W, Chen, X, Sprengeler, P.A, Zhang, X, Graber, A, Reich, S.H, Stopatschinskaja, S, Solomon, B, Besse, B, Drilon, A. | | Deposit date: | 2021-09-30 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Molecular Characteristics of Repotrectinib That Enable Potent Inhibition of TRK Fusion Proteins and Resistant Mutations.
Mol.Cancer Ther., 20, 2021
|
|
