8QD2
 
 | | Ayg1p in complex with 1,3-Dihydroxynaphthalene | | Descriptor: | Pigment biosynthesis protein yellowish-green 1, naphthalene-1,3-diol | | Authors: | Schmalhofer, M, Vagstad, A.L, Zhou, Q, Bode, H.B, Groll, M. | | Deposit date: | 2023-08-28 | | Release date: | 2024-03-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Polyketide Trimming Shapes Dihydroxynaphthalene-Melanin and Anthraquinone Pigments.
Adv Sci, 11, 2024
|
|
8EOO
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing human antibodies WRAIR-2063 and WRAIR-2151 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Spike protein S1, ... | | Authors: | Jensen, J.L, Sankhala, R.S, Joyce, M.G. | | Deposit date: | 2022-10-03 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Targeting the Spike Receptor Binding Domain Class V Cryptic Epitope by an Antibody with Pan-Sarbecovirus Activity.
J.Virol., 97, 2023
|
|
6WOK
 
 | | Crystal structure of estrogen receptor alpha in complex with receptor degrader 6 | | Descriptor: | (1R,3R)-1-(2,6-difluoro-4-{2-[3-(fluoromethyl)azetidin-1-yl]ethoxy}phenyl)-2-(2-fluoro-2-methylpropyl)-3-methyl-2,3,4,9-tetrahydro-1H-beta-carboline, (2S)-3-(3-hydroxyphenyl)-2-(4-iodophenyl)-4-methyl-2H-1-benzopyran-6-ol, Estrogen receptor | | Authors: | Kiefer, J.R, Vinogradova, M, Liang, J, Zhang, B, Wang, X, Labadie, S. | | Deposit date: | 2020-04-24 | | Release date: | 2020-07-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.309 Å) | | Cite: | Discovery of GNE-149 as a Full Antagonist and Efficient Degrader of Estrogen Receptor alpha for ER+ Breast Cancer.
Acs Med.Chem.Lett., 11, 2020
|
|
8QD4
 
 | | Ayg1p active site converted to tetrahedral sulfonate ester | | Descriptor: | Pigment biosynthesis protein yellowish-green 1, phenylmethanesulfonic acid | | Authors: | Schmalhofer, M, Vagstad, A.L, Zhou, Q, Bode, H.B, Groll, M. | | Deposit date: | 2023-08-28 | | Release date: | 2024-03-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Polyketide Trimming Shapes Dihydroxynaphthalene-Melanin and Anthraquinone Pigments.
Adv Sci, 11, 2024
|
|
7UGQ
 
 | |
7UPW
 
 | | Three RBD-down state of SARS-CoV-2 D614G spike in complex with the SP1-77 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Luo, S, Kreutzberger, A, Kirchhausen, T, Chen, B, Haynes, B, Alt, F. | | Deposit date: | 2022-04-18 | | Release date: | 2022-08-10 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | An antibody from single human V H -rearranging mouse neutralizes all SARS-CoV-2 variants through BA.5 by inhibiting membrane fusion.
Sci Immunol, 7, 2022
|
|
7UOH
 
 | | PRMT5/MEP50 crystal structure with MTA and an achiral, class 1, non-atropisomeric inhibitor bound | | Descriptor: | (2M)-2-[(4M)-4-{4-(aminomethyl)-1-oxo-8-[(2R)-oxolan-2-yl]-1,2-dihydrophthalazin-6-yl}-1-methyl-1H-pyrazol-5-yl]-1-benzothiophene-3-carbonitrile, 5'-DEOXY-5'-METHYLTHIOADENOSINE, Methylosome protein 50, ... | | Authors: | Gunn, R.J, Thomas, N.C, Lawson, J.D, Ivetac, A, Kulyk, S, Smith, C.R, Marx, M.A. | | Deposit date: | 2022-04-12 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design and evaluation of achiral, non-atropisomeric 4-(aminomethyl)phthalazin-1(2H)-one derivatives as novel PRMT5/MTA inhibitors.
Bioorg.Med.Chem., 71, 2022
|
|
8PWU
 
 | |
7UCG
 
 | |
8PE0
 
 | | X-ray structure of the Thermus thermophilus K167L mutant of the PilF-GSPIIB domain in the c-di-GMP bound state | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), ACETATE ION, SULFATE ION, ... | | Authors: | Neissner, K, Woehnert, J. | | Deposit date: | 2023-06-13 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structure of the K167L mutant of PilF-GSPIIB domain in the c-di-GMP bound state
To Be Published
|
|
8PK5
 
 | | INTS13-INTS14 complex with ZNF609 | | Descriptor: | Integrator complex subunit 13, Integrator complex subunit 14,Zinc finger protein 609, MAGNESIUM ION | | Authors: | Sabath, K, Jonas, S. | | Deposit date: | 2023-06-25 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Basis of gene-specific transcription regulation by the Integrator complex.
Mol.Cell, 84, 2024
|
|
8PI9
 
 | | DNA binding domain of HNF-1A bound to P2-HNF4A promoter DNA variant (P2 -181G>A) | | Descriptor: | Chains: E, Chains: F, Hepatocyte nuclear factor 1-alpha | | Authors: | Kind, L, Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2023-06-21 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular mechanism of HNF-1A-mediated HNF4A gene regulation and promoter-driven HNF4A-MODY diabetes.
JCI Insight, 9, 2024
|
|
6VRU
 
 | | PIM-inhibitor complex 1 | | Descriptor: | 3,4-dichloro-2-cyclopropyl-1-[(piperidin-4-yl)methyl]-1H-pyrrolo[2,3-b]pyridine-6-carboxamide, ACETATE ION, IMIDAZOLE, ... | | Authors: | Barberis, C.E, Batchelor, J.D, Mechin, I, Liu, J. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Discovery of SARxxxx92, a pan-PIM kinase inhibitor, efficacious in a KG1 tumor model.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
8HGU
 
 | |
6VIX
 
 | | BRD4_Bromodomain2 complex with pyrrolopyridone compound 18 | | Descriptor: | 4-[2-(2,4-difluorophenoxy)-5-(methylsulfonyl)phenyl]-N-ethyl-6-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridine-2-carboxamide, Bromodomain-containing protein 4 | | Authors: | Longenecker, K.L, Park, C.H, Qiu, W. | | Deposit date: | 2020-01-14 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.116 Å) | | Cite: | Discovery ofN-Ethyl-4-[2-(4-fluoro-2,6-dimethyl-phenoxy)-5-(1-hydroxy-1-methyl-ethyl)phenyl]-6-methyl-7-oxo-1H-pyrrolo[2,3-c]pyridine-2-carboxamide (ABBV-744), a BET Bromodomain Inhibitor with Selectivity for the Second Bromodomain.
J.Med.Chem., 63, 2020
|
|
8HMO
 
 | |
6VKC
 
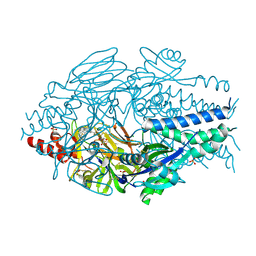 | | Crystal Structure of Inhibitor JNJ-36811054 in Complex with Prefusion RSV F Glycoprotein | | Descriptor: | 3-{[5-chloro-1-(4,4,4-trifluorobutyl)-1H-imidazo[4,5-b]pyridin-2-yl]methyl}-1-cyclopropyl-1,3-dihydro-2H-imidazo[4,5-c]pyridin-2-one, CHLORIDE ION, D(-)-TARTARIC ACID, ... | | Authors: | McLellan, J.S. | | Deposit date: | 2020-01-20 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of 3-({5-Chloro-1-[3-(methylsulfonyl)propyl]-1H-indol-2-yl}methyl)-1-(2,2,2-trifluoroethyl)-1,3-dihydro-2H-imidazo[4,5-c]pyridin-2-one (JNJ-53718678), a Potent and Orally Bioavailable Fusion Inhibitor of Respiratory Syncytial Virus.
J.Med.Chem., 63, 2020
|
|
6WMQ
 
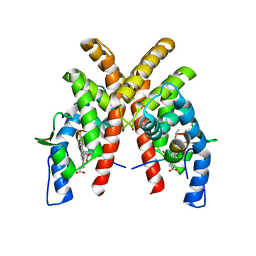 | |
6WYK
 
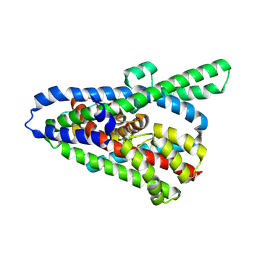 | | Cryo-EM structure of the GltPh L152C-G321C mutant in the intermediate chloride conducting state. | | Descriptor: | ASPARTIC ACID, Glutamate transporter homolog | | Authors: | Font, J, Chen, I, Sobti, M, Stewart, A.G, Ryan, R.M. | | Deposit date: | 2020-05-13 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Glutamate transporters have a chloride channel with two hydrophobic gates.
Nature, 591, 2021
|
|
8D12
 
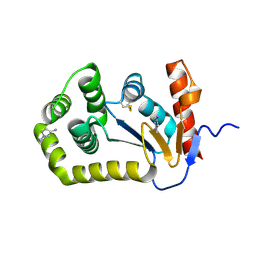 | |
8CZM
 
 | |
8DG0
 
 | |
8T2L
 
 | | Crystal structure of LC3A in complex with the LIR of NSs4 | | Descriptor: | Non-structural protein S,Microtubule-associated proteins 1A/1B light chain 3A chimera | | Authors: | Ali, M.G.H, Wahba, H.M, Cyr, N, Omichinski, J.G. | | Deposit date: | 2023-06-06 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | An LIR motif in the Rift Valley fever virus NSs protein is critical for the interaction with LC3 family members and inhibition of autophagy.
Plos Pathog., 20, 2024
|
|
8TGO
 
 | | Crystal structure of the BG505 triple tandem trimer gp140 HIV-1 Env in complex with PGT124 and 35O22 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35O22 scFv, ... | | Authors: | Xian, Y, Yuan, M, Wilson, I.A. | | Deposit date: | 2023-07-12 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (5.75 Å) | | Cite: | Triple tandem trimer immunogens for HIV-1 and influenza nucleic acid-based vaccines.
Npj Vaccines, 9, 2024
|
|
8CXD
 
 | |
