5EDC
 
 | |
6AGG
 
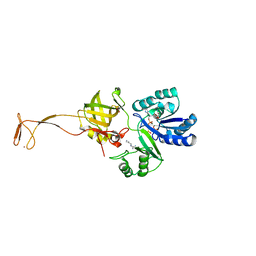 | |
2JDZ
 
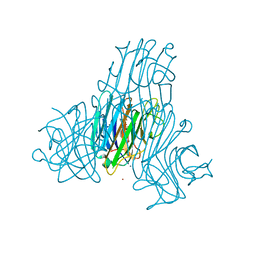 | | Crystal structure of recombinant Dioclea guianensis lectin complexed with 5-bromo-4-chloro-3-indolyl-a-D-mannose | | Descriptor: | 5-bromo-4-chloro-1H-indol-3-yl alpha-D-mannopyranoside, CADMIUM ION, CALCIUM ION, ... | | Authors: | Nagano, C.S, Sanz, L, Cavada, B.S, Calvete, J.J. | | Deposit date: | 2007-01-12 | | Release date: | 2007-10-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights Into the Structural Basis of the Ph-Dependent Dimer-Tetramer Equilibrium Through Crystallographic Analysis of Recombinant Diocleinae Lectins.
Biochem.J., 409, 2008
|
|
6TU5
 
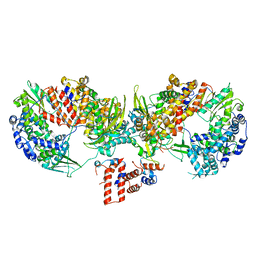 | | Influenza A/H7N9 polymerase core (apo) | | Descriptor: | MAGNESIUM ION, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Cusack, S, Pflug, A. | | Deposit date: | 2020-01-02 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.325 Å) | | Cite: | A Structure-Based Model for the Complete Transcription Cycle of Influenza Polymerase.
Cell, 181, 2020
|
|
8SQ9
 
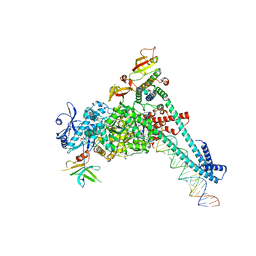 | | SARS-CoV-2 replication-transcription complex bound to nsp9 and UMPCPP, as a pre-catalytic NMPylation intermediate | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]uridine, MAGNESIUM ION, Non-structural protein 7, ... | | Authors: | Small, G.I, Darst, S.A, Campbell, E.A. | | Deposit date: | 2023-05-04 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and functional insights into the enzymatic plasticity of the SARS-CoV-2 NiRAN domain.
Mol.Cell, 83, 2023
|
|
6TGZ
 
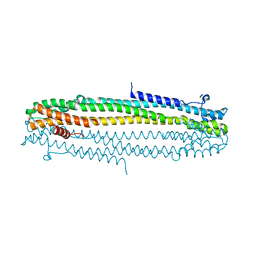 | | IE1 from human cytomegalovirus | | Descriptor: | 55 kDa immediate-early protein 1 | | Authors: | Schweininger, J, Muller, Y.A. | | Deposit date: | 2019-11-18 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Cytomegalovirus immediate-early 1 proteins form a structurally distinct protein class with adaptations determining cross-species barriers.
Plos Pathog., 17, 2021
|
|
5E0L
 
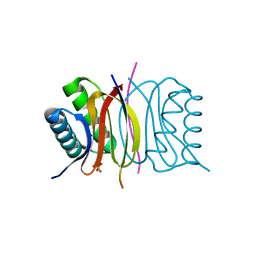 | | LC8 - Chica (415-424) Complex | | Descriptor: | Dynein light chain 1, cytoplasmic, Protein Chica peptide, ... | | Authors: | Clark, S.A, Barbar, E.B, Karplus, P.A. | | Deposit date: | 2015-09-29 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | The Anchored Flexibility Model in LC8 Motif Recognition: Insights from the Chica Complex.
Biochemistry, 55, 2016
|
|
6GU7
 
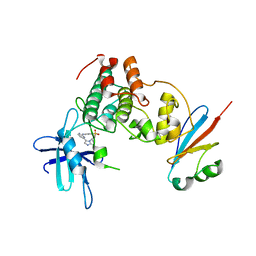 | | CDK1/Cks2 in complex with AZD5438 | | Descriptor: | 4-(2-methyl-3-propan-2-yl-imidazol-4-yl)-~{N}-(4-methylsulfonylphenyl)pyrimidin-2-amine, Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 2 | | Authors: | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | Deposit date: | 2018-06-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
8HST
 
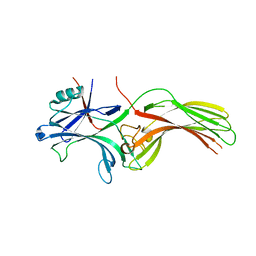 | |
8SQJ
 
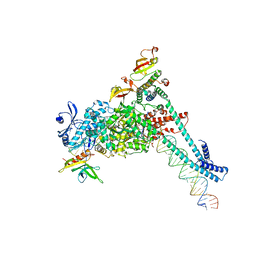 | | SARS-CoV-2 replication-transcription complex bound to RNA-nsp9, as a noncatalytic RNA-nsp9 binding mode | | Descriptor: | 5'-O-[(R)-hydroxy(thiophosphonooxy)phosphoryl]guanosine, MAGNESIUM ION, Non-structural protein 7, ... | | Authors: | Small, G.I, Darst, S.A, Campbell, E.A. | | Deposit date: | 2023-05-04 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural and functional insights into the enzymatic plasticity of the SARS-CoV-2 NiRAN domain.
Mol.Cell, 83, 2023
|
|
8I60
 
 | |
8HSV
 
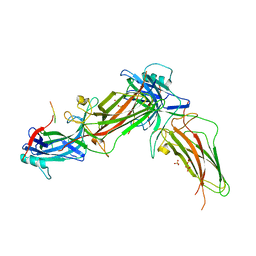 | | The structure of rat beta-arrestin1 in complex with a rat Mdm2 peptide | | Descriptor: | Beta-arrestin-1, SULFATE ION, peptide from E3 ubiquitin-protein ligase Mdm2 | | Authors: | Yun, Y, Yoon, H.J, Choi, Y, Lee, H.H. | | Deposit date: | 2022-12-20 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | GPCR targeting of E3 ubiquitin ligase MDM2 by inactive beta-arrestin.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8SQK
 
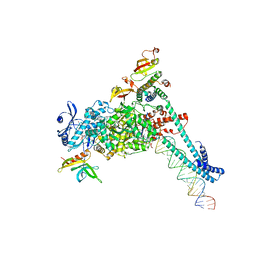 | | SARS-CoV-2 replication-transcription complex bound to RNA-nsp9 and GDP-betaS, as a pre-catalytic deRNAylation/mRNA capping intermediate | | Descriptor: | 5'-O-[(R)-hydroxy(thiophosphonooxy)phosphoryl]guanosine, MAGNESIUM ION, Non-structural protein 7, ... | | Authors: | Small, G.I, Darst, S.A, Campbell, E.A. | | Deposit date: | 2023-05-04 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural and functional insights into the enzymatic plasticity of the SARS-CoV-2 NiRAN domain.
Mol.Cell, 83, 2023
|
|
8T1J
 
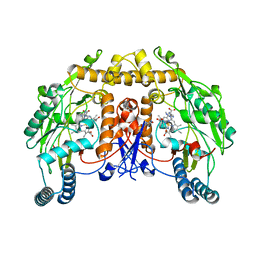 | | Uncrosslinked nNOS-CaM oxygenase homodimer | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ARGININE, Nitric oxide synthase 1, ... | | Authors: | Lee, K, Pospiech, T.H, Southworth, D. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mapping interactions of calmodulin and neuronal NO synthase by crosslinking and mass spectrometry.
J.Biol.Chem., 300, 2023
|
|
5ENG
 
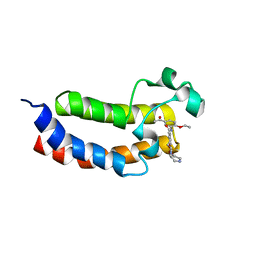 | | Crystal structure of the bromodomain of human CREBBP in complex with UP39 | | Descriptor: | CREB-binding protein, methyl 2-[2-(3,5-dihydro-2~{H}-pyrazin-4-yl)ethoxy]-5-[(5-ethanoyl-2-ethoxy-phenyl)carbamoyl]benzoate | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2015-11-09 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Binding Motifs in the CBP Bromodomain: An Analysis of 20 Crystal Structures of Complexes with Small Molecules.
ACS Med Chem Lett, 9, 2018
|
|
8IFF
 
 | | Cryo-EM structure of Arabidopsis phytochrome A. | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome A | | Authors: | Ma, L, Zhou, C, Wang, J, Guan, Z, Yin, P. | | Deposit date: | 2023-02-17 | | Release date: | 2023-08-02 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Plant phytochrome A in the Pr state assembles as an asymmetric dimer.
Cell Res., 33, 2023
|
|
2JE7
 
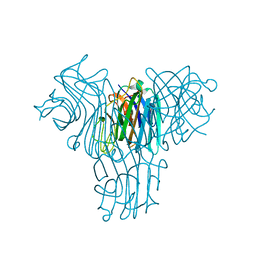 | | Crystal structure of recombinant Dioclea guianensis lectin S131H complexed with 5-bromo-4-chloro-3-indolyl-a-D-mannose | | Descriptor: | 5-bromo-4-chloro-1H-indol-3-yl alpha-D-mannopyranoside, CALCIUM ION, LECTIN ALPHA CHAIN, ... | | Authors: | Nagano, C.S, Sanz, L, Cavada, B.S, Calvete, J.J. | | Deposit date: | 2007-01-15 | | Release date: | 2007-10-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Insights Into the Structural Basis of the Ph- Dependent Dimer-Tetramer Equilibrium Through Crystallographic Analysis of Recombinant Diocleinae Lectins.
Biochem.J., 409, 2008
|
|
5EP1
 
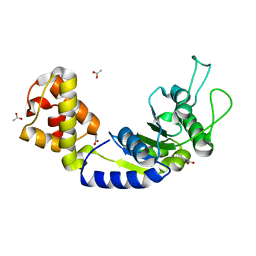 | | Quorum-Sensing Signal Integrator LuxO - Catalytic Domain | | Descriptor: | ACETATE ION, Putative repressor protein luxO | | Authors: | Shah, T, Selcuk, H.B, Jeffrey, P.D, Hughson, F.M. | | Deposit date: | 2015-11-11 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure, Regulation, and Inhibition of the Quorum-Sensing Signal Integrator LuxO.
Plos Biol., 14, 2016
|
|
8T1K
 
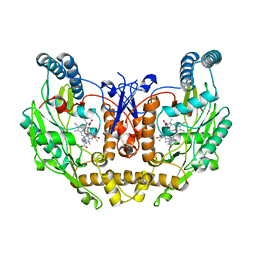 | | DSBU crosslinked nNOS-CaM oxygenase homodimer | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ARGININE, Nitric oxide synthase 1, ... | | Authors: | Lee, K, Pospiech, T.H, Southworth, D. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Mapping interactions of calmodulin and neuronal NO synthase by crosslinking and mass spectrometry.
J.Biol.Chem., 300, 2023
|
|
5EP7
 
 | |
5EPI
 
 | |
5EG2
 
 | | SET7/9 N265A in complex with AdoHcy and TAF10 peptide | | Descriptor: | Histone-lysine N-methyltransferase SETD7, S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION, ... | | Authors: | Kroner, G.M, Fick, R.J, Trievel, R.C. | | Deposit date: | 2015-10-26 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Sulfur-Oxygen Chalcogen Bonding Mediates AdoMet Recognition in the Lysine Methyltransferase SET7/9.
Acs Chem.Biol., 11, 2016
|
|
6NOW
 
 | |
5EPL
 
 | | Crystal Structure of chromodomain of CBX4 in complex with inhibitor UNC3866 | | Descriptor: | E3 SUMO-protein ligase CBX4, UNKNOWN ATOM OR ION, unc3866 | | Authors: | Liu, Y, Tempel, W, Walker, J.R, Stuckey, J.I, Dickson, B.M, James, L.I, Frye, S.V, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-11 | | Release date: | 2015-12-23 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | A cellular chemical probe targeting the chromodomains of Polycomb repressive complex 1.
Nat.Chem.Biol., 12, 2016
|
|
3EED
 
 | | Crystal structure of human protein kinase CK2 regulatory subunit (CK2beta; mutant 1-193) | | Descriptor: | Casein kinase II subunit beta, SULFATE ION, ZINC ION | | Authors: | Niefind, K, Raaf, J, Issinger, O.-G. | | Deposit date: | 2008-09-04 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The interaction of CK2{alpha} and CK2{beta}, the subunits of protein kinase CK2, requires CK2{beta} in a preformed conformation and is enthalpically driven
Protein Sci., 17, 2008
|
|
