2XB2
 
 | | Crystal structure of the core Mago-Y14-eIF4AIII-Barentsz-UPF3b assembly shows how the EJC is bridged to the NMD machinery | | Descriptor: | EUKARYOTIC INITIATION FACTOR 4A-III, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Buchwald, G, Ebert, J, Basquin, C, Sauliere, J, Jayachandran, U, Bono, F, Le Hir, H, Conti, E. | | Deposit date: | 2010-04-03 | | Release date: | 2010-05-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Insights Into the Recruitment of the Nmd Machinery from the Crystal Structure of a Core Ejc-Upf3B Complex.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7K0Y
 
 | | Cryo-EM structure of activated-form DNA-PK (complex VI) | | Descriptor: | DNA (5'-D(*AP*AP*GP*CP*AP*GP*TP*AP*GP*AP*GP*CP*A)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*CP*TP*CP*TP*AP*CP*TP*GP*CP*TP*TP*CP*GP*AP*TP*AP*TP*CP*G)-3'), DNA-dependent protein kinase catalytic subunit, ... | | Authors: | Chen, X, Gellert, M, Yang, W. | | Deposit date: | 2020-09-06 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of an activated DNA-PK and its implications for NHEJ.
Mol.Cell, 81, 2021
|
|
7K1J
 
 | | CryoEM structure of inactivated-form DNA-PK (Complex III) | | Descriptor: | DNA (5'-D(*AP*AP*GP*CP*AP*GP*TP*AP*GP*AP*GP*CP*A)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*CP*TP*CP*TP*AP*CP*TP*GP*CP*TP*TP*CP*GP*AP*TP*AP*TP*CP*G)-3'), DNA-dependent protein kinase catalytic subunit, ... | | Authors: | Chen, X, Gellert, M, Yang, W. | | Deposit date: | 2020-09-07 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of an activated DNA-PK and its implications for NHEJ.
Mol.Cell, 81, 2021
|
|
6D90
 
 | | Mammalian 80S ribosome with a double translocated CrPV-IRES, P-site tRNA and eRF1. | | Descriptor: | 18S rRNA, 28S rRNA, 5.8S rRNA, ... | | Authors: | Pisareva, V.P, Pisarev, A.V, Fernandez, I.S. | | Deposit date: | 2018-04-27 | | Release date: | 2018-06-06 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Dual tRNA mimicry in the Cricket Paralysis Virus IRES uncovers an unexpected similarity with the Hepatitis C Virus IRES.
Elife, 7, 2018
|
|
7K1N
 
 | | CryoEM structure of inactivated-form DNA-PK (Complex V) | | Descriptor: | DNA (5'-D(P*AP*AP*GP*CP*AP*GP*TP*AP*GP*AP*GP*CP*A)-3'), DNA (5'-D(P*GP*CP*AP*TP*GP*CP*TP*CP*TP*AP*CP*TP*GP*CP*TP*TP*CP*GP*AP*TP*AP*TP*CP*G)-3'), DNA-dependent protein kinase catalytic subunit, ... | | Authors: | Chen, X, Gellert, M, Yang, W. | | Deposit date: | 2020-09-08 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of an activated DNA-PK and its implications for NHEJ.
Mol.Cell, 81, 2021
|
|
8K2C
 
 | | Cryo-EM structure of the human 80S ribosome with Tigecycline | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Li, X, Wang, M, Cheng, J. | | Deposit date: | 2023-07-12 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
7K1K
 
 | | CryoEM structure of inactivated-form DNA-PK (Complex IV) | | Descriptor: | DNA (5'-D(*AP*AP*GP*CP*AP*GP*TP*AP*GP*AP*GP*CP*A)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*CP*TP*CP*TP*AP*CP*TP*GP*CP*TP*TP*CP*GP*AP*TP*AP*TP*CP*G)-3'), DNA-dependent protein kinase catalytic subunit, ... | | Authors: | Chen, X, Gellert, M, Yang, W. | | Deposit date: | 2020-09-07 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of an activated DNA-PK and its implications for NHEJ.
Mol.Cell, 81, 2021
|
|
6QB8
 
 | | Human CCT:mLST8 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, ... | | Authors: | Cuellar, J, Santiago, C, Ludlam, W.G, Bueno-Carrasco, M.T, Valpuesta, J.M, Willardson, B.M. | | Deposit date: | 2018-12-20 | | Release date: | 2019-07-03 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Structural and functional analysis of the role of the chaperonin CCT in mTOR complex assembly.
Nat Commun, 10, 2019
|
|
2XWU
 
 | | CRYSTAL STRUCTURE OF IMPORTIN 13 - UBC9 COMPLEX | | Descriptor: | IMPORTIN13, SUMO-CONJUGATING ENZYME UBC9 | | Authors: | Gruenwald, M, Bono, F. | | Deposit date: | 2010-11-05 | | Release date: | 2010-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Importin13-Ubc9 Complex: Nuclear Import and Release of a Key Regulator of Sumoylation.
Embo J., 30, 2011
|
|
8IUE
 
 | | RNA polymerase III pre-initiation complex melting complex 1 | | Descriptor: | DNA (74-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Hou, H, Jin, Q, Ren, Y, Wang, Q, Xu, Y. | | Deposit date: | 2023-03-24 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of the SNAPc-bound RNA polymerase III preinitiation complex.
Cell Res., 33, 2023
|
|
8OSL
 
 | | Cryo-EM structure of CLOCK-BMAL1 bound to the native Por enhancer nucleosome (map 2, additional 3D classification and flexible refinement) | | Descriptor: | Basic helix-loop-helix ARNT-like protein 1, Circadian locomoter output cycles protein kaput, DNA (147-MER), ... | | Authors: | Michael, A.K, Stoos, L, Kempf, G, Cavadini, S, Thoma, N. | | Deposit date: | 2023-04-19 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
8OSK
 
 | | Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL+5.8 (composite map) | | Descriptor: | Basic helix-loop-helix ARNT-like protein 1, Circadian locomoter output cycles protein kaput, DNA (124-MER), ... | | Authors: | Stoos, L, Michael, A.K, Kempf, G, Cavadini, S, Thoma, N.H. | | Deposit date: | 2023-04-19 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
8OSJ
 
 | | Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL-6.2 (DNA conformation 1) | | Descriptor: | Basic helix-loop-helix ARNT-like protein 1, Circadian locomoter output cycles protein kaput, DNA (124-MER), ... | | Authors: | Michael, A.K, Stoos, L, Kempf, G, Cavadini, S, Thoma, N.H. | | Deposit date: | 2023-04-19 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Cooperation between bHLH transcription factors and histones for DNA access.
Nature, 619, 2023
|
|
8I0P
 
 | | The cryo-EM structure of human pre-Bact complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Cell division cycle 5-like protein, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2023-01-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis for the activation of human spliceosome.
Nat Commun, 15, 2024
|
|
7YWX
 
 | | Structure of the human CCAN CENP-A alpha-satellite complex | | Descriptor: | Centromere protein C, Centromere protein H, Centromere protein I, ... | | Authors: | Yatskevich, S, Muir, K.W, Bellini, D, Zhang, Z, Yang, J, Tischer, T, Predin, M, Dendooven, T, McLaughlin, S.H, Barford, D. | | Deposit date: | 2022-02-14 | | Release date: | 2022-05-18 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structure of the human inner kinetochore bound to a centromeric CENP-A nucleosome.
Science, 376, 2022
|
|
7ZDD
 
 | | Crystal structure of TRIM33 PHD-Bromodomain isoform B in complex with H3K10ac histone peptide. | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase TRIM33, Histone H3.X, ... | | Authors: | Caria, S, Duclos, S, Crespillo, S, Errey, J, Barker, J.J. | | Deposit date: | 2022-03-29 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.625 Å) | | Cite: | Identification of Histone Peptide Binding Specificity and Small-Molecule Ligands for the TRIM33 alpha and TRIM33 beta Bromodomains.
Acs Chem.Biol., 17, 2022
|
|
7Z21
 
 | | BAF A12T bound to the lamin A/C Ig-fold domain | | Descriptor: | Barrier-to-autointegration factor, N-terminally processed, CHLORIDE ION, ... | | Authors: | Marcelot, A, Legrand, P, Zinn-Justin, S. | | Deposit date: | 2022-02-25 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.629 Å) | | Cite: | The BAF A12T mutation disrupts lamin A/C interaction, impairing robust repair of nuclear envelope ruptures in Nestor-Guillermo progeria syndrome cells.
Nucleic Acids Res., 50, 2022
|
|
7KW7
 
 | | Atomic cryoEM structure of Hsp90-Hsp70-Hop-GR | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glucocorticoid receptor, Heat shock 70 kDa protein 1A, ... | | Authors: | Wang, R.Y, Noddings, C.M, Kirschke, E, Myasnikov, A, Johnson, J.L, Agard, D.A. | | Deposit date: | 2020-11-30 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structure of Hsp90-Hsp70-Hop-GR reveals the Hsp90 client-loading mechanism.
Nature, 601, 2022
|
|
3A4S
 
 | | The crystal structure of the SLD2:Ubc9 complex | | Descriptor: | NFATC2-interacting protein, SUMO-conjugating enzyme UBC9 | | Authors: | Sekiyama, N, Arita, K, Ikeda, Y, Ariyoshi, M, Tochio, H, Saitoh, H, Shirakawa, M. | | Deposit date: | 2009-07-14 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for regulation of poly-SUMO chain by a SUMO-like domain of Nip45
Proteins, 78, 2009
|
|
7R5R
 
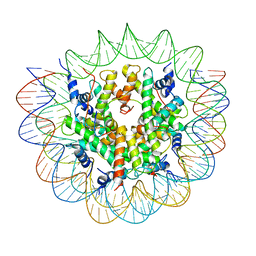 | | Structure of the human CCAN CENP-A alpha-satellite complex | | Descriptor: | Centromere protein C, DNA (171-MER), Histone H2A type 1-C, ... | | Authors: | Yatskevich, S, Muir, K.W, Bellini, D, Zhang, Z, Yang, J, Tischer, T, Predin, M, Dendooven, T, McLaughlin, S.H, Barford, D. | | Deposit date: | 2022-02-11 | | Release date: | 2022-04-27 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Structure of the human inner kinetochore bound to a centromeric CENP-A nucleosome.
Science, 376, 2022
|
|
7QGG
 
 | | Neuronal RNA granules are ribosome complexes stalled at the pre-translocation state | | Descriptor: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Pulk, A, Kipper, K, Mansour, A. | | Deposit date: | 2021-12-08 | | Release date: | 2022-10-26 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Neuronal RNA granules are ribosome complexes stalled at the pre-translocation state.
J.Mol.Biol., 434, 2022
|
|
8FKY
 
 | |
8FLA
 
 | |
8FLD
 
 | |
8HNI
 
 | | hnRNP A2/B1 RRMs in complex with telomeric DNA | | Descriptor: | DNA (5'-D(P*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*T)-3'), Heterogeneous nuclear ribonucleoproteins A2/B1 | | Authors: | Liu, Y, Abula, A, Xiao, H, Guo, H, Li, T, Zheng, L, Chen, B, Nguyen, H, Ji, X. | | Deposit date: | 2022-12-07 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.644 Å) | | Cite: | Structural Insight Into hnRNP A2/B1 Homodimerization and DNA Recognition.
J.Mol.Biol., 435, 2023
|
|
