5MTR
 
 | | Crystal structure of M. tuberculosis InhA inhibited by PT512 | | Descriptor: | 2-[4-[(4-cyclopentyl-1,2,3-triazol-1-yl)methyl]-2-oxidanyl-phenoxy]benzenecarbonitrile, CHLORIDE ION, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Eltschkner, S, Pschibul, A, Spagnuolo, L.A, Yu, W, Tonge, P.J, Kisker, C. | | Deposit date: | 2017-01-10 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evaluating the Contribution of Transition-State Destabilization to Changes in the Residence Time of Triazole-Based InhA Inhibitors.
J. Am. Chem. Soc., 139, 2017
|
|
5MUR
 
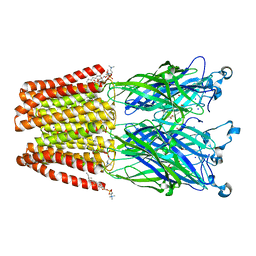 | | X-ray structure of the F14'A mutant of GLIC in complex with propofol | | Descriptor: | 2,6-BIS(1-METHYLETHYL)PHENOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Sauguet, L, Fourati, Z, Delarue, M. | | Deposit date: | 2017-01-13 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis for a Bimodal Allosteric Mechanism of General Anesthetic Modulation in Pentameric Ligand-Gated Ion Channels.
Cell Rep, 23, 2018
|
|
6RGZ
 
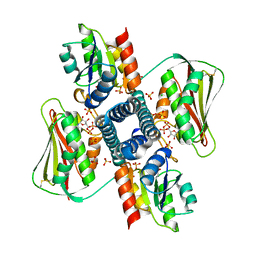 | | Revisiting pH-gated conformational switch. Complex HK853-RR468 pH 6.5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-18 | | Release date: | 2020-02-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
6RH8
 
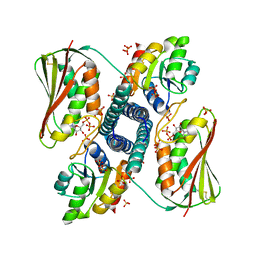 | | Revisiting pH-gated conformational switch. Complex HK853 mutant H260A -RR468 mutant D53A pH 5.3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Response regulator, SULFATE ION, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-19 | | Release date: | 2020-02-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
5MGA
 
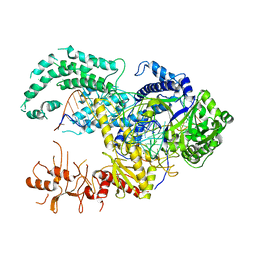 | |
5M60
 
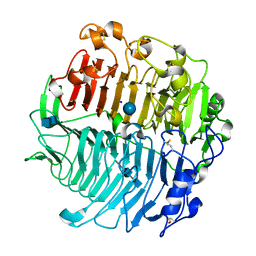 | | Chaetomium thermophilum beta-1-3-glucanase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-1,3-glucanase, SODIUM ION, ... | | Authors: | Papageorgiou, A.C, Chen, J, Li, D. | | Deposit date: | 2016-10-23 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure and biological implications of a glycoside hydrolase family 55 beta-1,3-glucanase from Chaetomium thermophilum.
Biochim. Biophys. Acta, 1865, 2017
|
|
6RLV
 
 | | Trypanosoma brucei Seryl-tRNA Synthetase in Complex with 5'-O-(N-(L-seryl)-sulfamoyl)N3-methyluridine | | Descriptor: | 5'-O-(N-(L-seryl)-sulfamoyl)N3-methyluridine, GLYCEROL, MALONATE ION, ... | | Authors: | Pang, L, De Graef, S, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2019-05-02 | | Release date: | 2020-01-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into the Binding of Natural Pyrimidine-Based Inhibitors of Class II Aminoacyl-tRNA Synthetases.
Acs Chem.Biol., 15, 2020
|
|
5M72
 
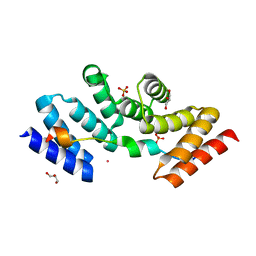 | | Structure of the human SRP68-72 protein-binding domain complex | | Descriptor: | GLYCEROL, POTASSIUM ION, SULFATE ION, ... | | Authors: | Becker, M.M.M, Wild, K, Sinning, I. | | Deposit date: | 2016-10-26 | | Release date: | 2016-12-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of human SRP72 complexes provide insights into SRP RNA remodeling and ribosome interaction.
Nucleic Acids Res., 45, 2017
|
|
6RML
 
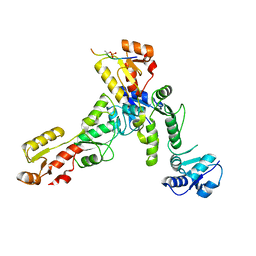 | | Crystal structure of TOPBP1 BRCT0,1,2 in complex with a 53BP1 phosphopeptide | | Descriptor: | 53BP1, DNA topoisomerase 2-binding protein 1 | | Authors: | Day, M, Oliver, A.W, Pearl, L.H. | | Deposit date: | 2019-05-07 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Phosphorylation-mediated interactions with TOPBP1 couple 53BP1 and 9-1-1 to control the G1 DNA damage checkpoint.
Elife, 8, 2019
|
|
6RMS
 
 | |
6RJ3
 
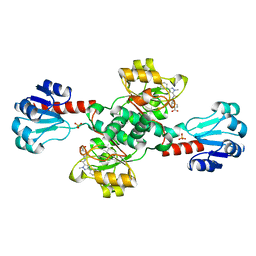 | | Crystal structure of PHGDH in complex with compound 15 | | Descriptor: | 4-[(1~{R})-1-[(2-methyl-5-phenyl-pyrazol-3-yl)carbonylamino]ethyl]benzoic acid, D-3-phosphoglycerate dehydrogenase, SULFATE ION | | Authors: | Bader, G, Wolkerstorfer, B, Zoephel, A. | | Deposit date: | 2019-04-26 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Intracellular Trapping of the Selective Phosphoglycerate Dehydrogenase (PHGDH) InhibitorBI-4924Disrupts Serine Biosynthesis.
J.Med.Chem., 62, 2019
|
|
5MKG
 
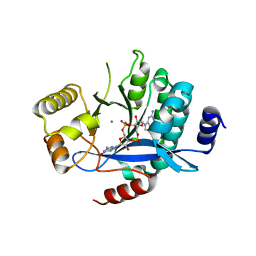 | | PA3825-EAL Ca-CdG Structure | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CALCIUM ION, Diguanylate phosphodiesterase | | Authors: | Horrell, S, Bellini, D, Strange, R, Wagner, A, Walsh, M. | | Deposit date: | 2016-12-04 | | Release date: | 2016-12-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Dimerisation induced formation of the active site and the identification of three metal sites in EAL-phosphodiesterases.
Sci Rep, 7, 2017
|
|
6RK8
 
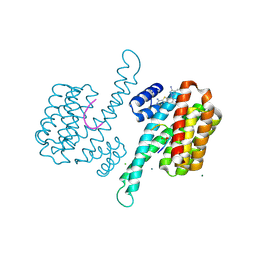 | | Fragment AZ-014 binding at the p53pT387/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 7-(3-azanyl-4-methyl-pyrazol-1-yl)-1-benzothiophene-2-carboximidamide, CHLORIDE ION, ... | | Authors: | Genet, S, Wolter, M, Guillory, X, Somsen, B, Leysen, S, Patel, J, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-04-30 | | Release date: | 2020-06-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
5M7C
 
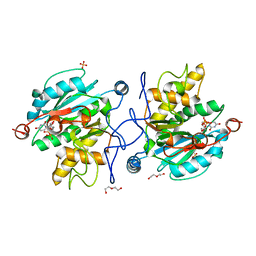 | |
5M9G
 
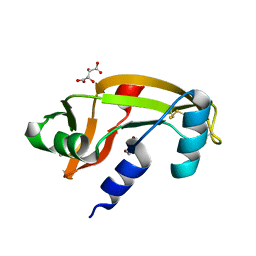 | | Human angiogenin PD variant K54R | | Descriptor: | Angiogenin, D(-)-TARTARIC ACID, DI(HYDROXYETHYL)ETHER | | Authors: | Bradshaw, W.J, Rehman, S, Pham, T.T.K, Thiyagarajan, N, Lee, R.L, Subramanian, V, Acharya, K.R. | | Deposit date: | 2016-11-01 | | Release date: | 2017-02-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural insights into human angiogenin variants implicated in Parkinson's disease and Amyotrophic Lateral Sclerosis.
Sci Rep, 7, 2017
|
|
5MLN
 
 | |
6ROR
 
 | | REP related 18-mer DNA | | Descriptor: | REP related 18-mer DNA from C. hominis, STRONTIUM ION | | Authors: | Kolenko, P, Svoboda, J, Schneider, B. | | Deposit date: | 2019-05-13 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural variability of CG-rich DNA 18-mers accommodating double T-T mismatches.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
5MLU
 
 | | Crystal structure of the PFV GAG CBS bound to a mononucleosome | | Descriptor: | DNA (145-MER), Histone H2A type 1, Histone H2B, ... | | Authors: | Pye, V.E, Maskell, D.P, Lesbats, P, Cherepanov, P. | | Deposit date: | 2016-12-07 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for spumavirus GAG tethering to chromatin.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6RL3
 
 | | Fragment AZ-003 binding at the p53pT387/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 5-azanyl-4-phenyl-thiophene-2-carboximidamide, CHLORIDE ION, ... | | Authors: | Genet, S, Wolter, M, Guillory, X, Somsen, B, Leysen, S, Patel, J, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-05-01 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
5MMY
 
 | | Crystal structure of OXA10 with HEPES | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-lactamase OXA-10, ... | | Authors: | Brem, J. | | Deposit date: | 2016-12-12 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | (13)C-Carbamylation as a mechanistic probe for the inhibition of class D beta-lactamases by avibactam and halide ions.
Org. Biomol. Chem., 15, 2017
|
|
6RL6
 
 | | Fragment AZ-024 binding at the p53pT387/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, Cellular tumor antigen p53, ... | | Authors: | Genet, S, Wolter, M, Guillory, X, Somsen, B, Leysen, S, Patel, J, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-05-01 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
5MAV
 
 | |
5MOA
 
 | | ABA RECEPTOR FROM TOMATO, SlPYL1 | | Descriptor: | SlPYL1 | | Authors: | Moreno-Alvero, M, Yunta, C, Gonzalez-Guzman, M, Arbona, V, Granell, A, Martinez-Ripoll, M, Infantes, L, Rodriguez, P.L, Albert, A. | | Deposit date: | 2016-12-14 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of Ligand-Bound Intermediates of Crop ABA Receptors Highlights PP2C as Necessary ABA Co-receptor.
Mol Plant, 10, 2017
|
|
5MOC
 
 | | Crystal structure of 14-3-3sigma and a p53 C-terminal 12-mer synthetic phosphopeptide | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Andrei, S, Ottmann, C, Leysen, S. | | Deposit date: | 2016-12-14 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Small-molecule stabilization of the p53 - 14-3-3 protein-protein interaction.
FEBS Lett., 591, 2017
|
|
5MOL
 
 | | Human IgE-Fc crystal structure | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-ETHOXYETHOXY)ETHANOL, 2-(2-METHOXYETHOXY)ETHANOL, ... | | Authors: | Dore, K.A, Davies, A.M, Drinkwater, N, Beavil, A.J, McDonnell, J.M, Sutton, B.J. | | Deposit date: | 2016-12-14 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Thermal sensitivity and flexibility of the C epsilon 3 domains in immunoglobulin E.
Biochim. Biophys. Acta, 1865, 2017
|
|
