1T1R
 
 | | Crystal Structure of the Reductoisomerase Complexed with a Bisphosphonate | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, SULFATE ION, [(ISOQUINOLIN-1-YLAMINO)-PHOSPHONO-METHYL]-PHOSPHONIC ACID | | Authors: | Yajima, S, Hara, K, Sanders, J.M, Yin, F, Ohsawa, K, Wiesner, J, Jomaa, H, Oldfield, E. | | Deposit date: | 2004-04-17 | | Release date: | 2004-09-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic Structures of Two Bisphosphonate:1-Deoxyxylulose-5-Phosphate Reductoisomerase Complexes
J.Am.Chem.Soc., 126, 2004
|
|
4K35
 
 | | The structure of a glycoside hydrolase family 81 endo-[beta]-1,3-glucanase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, glycoside hydrolase family 81 endo-beta-1,3-glucanase | | Authors: | Jiang, Z.Q, Zhou, P, Chen, Z.Z, Yan, Q.J, Yang, S.Q, Hilgenfeld, R. | | Deposit date: | 2013-04-10 | | Release date: | 2013-10-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | The structure of a glycoside hydrolase family 81
endo-[beta]-1,3-glucanase
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4KB9
 
 | | Crystal structure of wild-type HIV-1 protease with novel tricyclic P2-ligands GRL-0739A | | Descriptor: | (3aR,3bR,4S,7aR,8aS)-decahydrofuro[2,3-b][1]benzofuran-4-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2013-04-23 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Highly Potent HIV-1 Protease Inhibitors with Novel Tricyclic P2 Ligands: Design, Synthesis, and Protein-Ligand X-ray Studies.
J.Med.Chem., 56, 2013
|
|
2F6Z
 
 | | Protein tyrosine phosphatase 1B with sulfamic acid inhibitors | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Tyrosine-protein phosphatase, ... | | Authors: | Evdokimov, A.G, Pokross, M.E, Klopfenstein, S.R. | | Deposit date: | 2005-11-29 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1,2,3,4-Tetrahydroisoquinolinyl sulfamic acids as phosphatase PTP1B inhibitors
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2O4N
 
 | | Crystal Structure of HIV-1 Protease (TRM Mutant) in Complex with Tipranavir | | Descriptor: | GLYCEROL, N-(3-{(1R)-1-[(6R)-4-HYDROXY-2-OXO-6-PHENETHYL-6-PROPYL-5,6-DIHYDRO-2H-PYRAN-3-YL]PROPYL}PHENYL)-5-(TRIFLUOROMETHYL)-2-PYRIDINESULFONAMIDE, protease | | Authors: | Kang, L.W, Armstrong, A.A, Muzammil, S, Jakalian, A, Bonneau, P.R, Schmelmer, V, Freire, E, Amzel, L.M. | | Deposit date: | 2006-12-04 | | Release date: | 2006-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Unique thermodynamic response of tipranavir to human immunodeficiency virus type 1 protease drug resistance mutations.
J.Virol., 81, 2007
|
|
1IR2
 
 | | Crystal Structure of Activated Ribulose-1,5-bisphosphate Carboxylase/oxygenase (Rubisco) from Green alga, Chlamydomonas reinhardtii Complexed with 2-Carboxyarabinitol-1,5-bisphosphate (2-CABP) | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, GLYCEROL, Large subunit of Rubisco, ... | | Authors: | Mizohata, E, Matsumura, H, Okano, Y, Kumei, M, Takuma, H, Onodera, J, Kato, K, Shibata, N, Inoue, T, Yokota, A, Kai, Y. | | Deposit date: | 2001-09-03 | | Release date: | 2002-03-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of activated ribulose-1,5-bisphosphate carboxylase/oxygenase from green alga Chlamydomonas reinhardtii complexed with 2-carboxyarabinitol-1,5-bisphosphate.
J.Mol.Biol., 316, 2002
|
|
2OO0
 
 | | A structural insight into the inhibition of human and Leishmania donovani ornithine decarboxylases by 3-aminooxy-1-aminopropane | | Descriptor: | 3-AMINOOXY-1-AMINOPROPANE, ACETATE ION, Ornithine decarboxylase, ... | | Authors: | Dufe, V.T, Ingner, D, Khomutov, A.R, Heby, O, Persson, L, Al-Karadaghi, S. | | Deposit date: | 2007-01-25 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A structural insight into the inhibition of human and Leishmania donovani ornithine decarboxylases by 1-amino-oxy-3-aminopropane.
Biochem.J., 405, 2007
|
|
2Q54
 
 | | Crystal structure of KB73 bound to HIV-1 protease | | Descriptor: | ACETATE ION, N~2~-ACETYL-N-[(1S,3S,4S)-4-({[(5S)-3-(3-ACETYLPHENYL)-2-OXO-1,3-OXAZOLIDIN-5-YL]CARBONYL}AMINO)-1-BENZYL-3-HYDROXY-5-PHENYLPENTYL]-L-VALINAMIDE, PHOSPHATE ION, ... | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2007-05-31 | | Release date: | 2007-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design and Synthesis of HIV-1 Protease Inhibitors Incorporating Oxazolidinones as P2/P2' Ligands in Pseudosymmetric Dipeptide Isosteres.
J.Med.Chem., 50, 2007
|
|
1T1S
 
 | | Crystal Structure of the Reductoisomerase Complexed with a Bisphosphonate | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Yajima, S, Hara, K, Sanders, J.M, Yin, F, Ohsawa, K, Wiesner, J, Jomaa, H, Oldfield, E. | | Deposit date: | 2004-04-17 | | Release date: | 2004-09-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic Structures of Two Bisphosphonate:1-Deoxyxylulose-5-Phosphate Reductoisomerase Complexes
J.Am.Chem.Soc., 126, 2004
|
|
4J55
 
 | | Crystal Structure of Multidrug Resistant HIV-1 Protease Clinical isolate PR20 with the potent antiviral inhibitor GRL-02031 | | Descriptor: | (3aS,5R,6aR)-hexahydro-2H-cyclopenta[b]furan-5-yl [(1S,2R)-1-benzyl-2-hydroxy-3-([(4-methoxyphenyl)sulfonyl]{[(2R)-5-oxopyrrolidin-2-yl]methyl}amino)propyl]carbamate, Protease | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2013-02-07 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Extreme Multidrug Resistant HIV-1 Protease with 20 Mutations Is Resistant to Novel Protease Inhibitors with P1'-Pyrrolidinone or P2-Tris-tetrahydrofuran.
J.Med.Chem., 56, 2013
|
|
2Q5K
 
 | | Crystal structure of lopinavir bound to wild type HIV-1 protease | | Descriptor: | N-{1-BENZYL-4-[2-(2,6-DIMETHYL-PHENOXY)-ACETYLAMINO]-3-HYDROXY-5-PHENYL-PENTYL}-3-METHYL-2-(2-OXO-TETRAHYDRO-PYRIMIDIN-1-YL)-BUTYRAMIDE, PHOSPHATE ION, Protease | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2007-06-01 | | Release date: | 2007-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design and Synthesis of HIV-1 Protease Inhibitors Incorporating Oxazolidinones as P2/P2' Ligands in Pseudosymmetric Dipeptide Isosteres.
J.Med.Chem., 50, 2007
|
|
4J54
 
 | | Crystal Structure of Multidrug Resistant HIV-1 Protease Clinical isolate PR20 with the potent antiviral inhibitor GRL-0519A | | Descriptor: | (3R,3aS,3bR,6aS,7aS)-octahydrodifuro[2,3-b:3',2'-d]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}propyl]carbamate, ACETATE ION, IODIDE ION, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2013-02-07 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Extreme Multidrug Resistant HIV-1 Protease with 20 Mutations Is Resistant to Novel Protease Inhibitors with P1'-Pyrrolidinone or P2-Tris-tetrahydrofuran.
J.Med.Chem., 56, 2013
|
|
2H3Z
 
 | | Structure of the HIV-1 matrix protein bound to di-C4-phosphatidylinositol-(4,5)-bisphosphate | | Descriptor: | (2R)-3-{[(R)-HYDROXY{[(1R,2R,3S,4R,5R,6S)-2,3,6-TRIHYDROXY-4,5-BIS(PHOSPHONOOXY)CYCLOHEXYL]OXY}PHOSPHORYL]OXY}PROPANE-1 ,2-DIYL DIBUTANOATE, Gag polyprotein | | Authors: | Saad, J.S, Miller, J, Tai, J, Kim, A, Ghanam, R.H, Summers, M.F. | | Deposit date: | 2006-05-23 | | Release date: | 2006-07-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1K5M
 
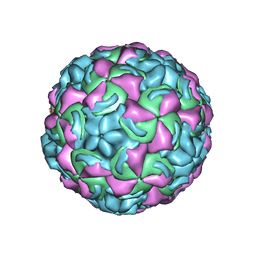 | | Crystal Structure of a Human Rhinovirus Type 14:Human Immunodeficiency Virus Type 1 V3 Loop Chimeric Virus MN-III-2 | | Descriptor: | CHIMERA OF HRV14 COAT PROTEIN VP2 (P1B) AND the V3 loop of HIV-1 gp120, COAT PROTEIN VP1 (P1D), COAT PROTEIN VP3 (P1C), ... | | Authors: | Ding, J, Smith, A.D, Geisler, S.C, Ma, X, Arnold, G.F, Arnold, E. | | Deposit date: | 2001-10-11 | | Release date: | 2002-07-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of a Human Rhinovirus
that Displays Part of the HIV-1 V3 Loop and
Induces Neutralizing Antibodies against
HIV-1
Structure, 10, 2002
|
|
4EE3
 
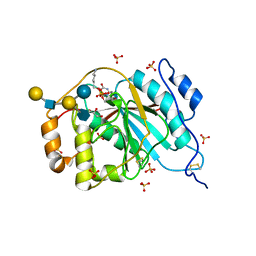 | | Crystal structure of human M340H-beta-1,4-galactosyltransferase-1 (M340H-B4GAL-T1) in complex with pentasaccharide | | Descriptor: | 6-AMINOHEXYL-URIDINE-C1,5'-DIPHOSPHATE, Beta-1,4-galactosyltransferase 1, GLYCEROL, ... | | Authors: | Ramakrishnan, B, Qasba, P.K. | | Deposit date: | 2012-03-28 | | Release date: | 2012-07-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Binding of N-acetylglucosamine (GlcNAc) beta 1-6-branched oligosaccharide acceptors to beta 4-galactosyltransferase I reveals a new ligand binding mode.
J.Biol.Chem., 287, 2012
|
|
2E40
 
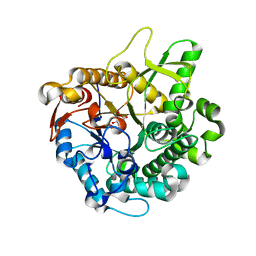 | | Crystal structure of intracellular family 1 beta-glucosidase BGL1A from the basidiomycete Phanerochaete chrysosporium in complex with gluconolactone | | Descriptor: | Beta-glucosidase, D-glucono-1,5-lactone | | Authors: | Nijikken, Y, Tsukada, T, Igarashi, K, Samejima, M, Fushinobu, S. | | Deposit date: | 2006-12-01 | | Release date: | 2007-03-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of intracellular family 1 beta-glucosidase BGL1A from the basidiomycete Phanerochaete chrysosporium
Febs Lett., 581, 2007
|
|
3R01
 
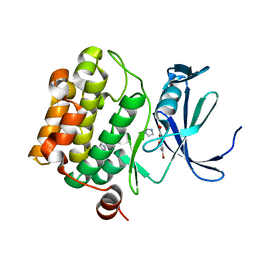 | | The discovery of novel benzofuran-2-carboxylic acids as potent Pim-1 inhibitors | | Descriptor: | 5-bromo-7-methoxy-1-benzofuran-2-carboxylic acid, IMIDAZOLE, Proto-oncogene serine/threonine-protein kinase pim-1 | | Authors: | Liu, J. | | Deposit date: | 2011-03-07 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The discovery of novel benzofuran-2-carboxylic acids as potent Pim-1 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
1XKX
 
 | | Kinetic and crystallographic studies on 2-(beta-D-glucopyranosyl)-5-methyl-1,3,4-oxadiazole,-benzothiazole, and-benzimidazole, inhibitors of muscle glycogen phosphorylase b. Evidence for a new binding site. | | Descriptor: | 2-(BETA-D-GLUCOPYRANOSYL)-5-METHYL-1-BENZIMIDAZOLE, Glycogen phosphorylase, muscle form, ... | | Authors: | Chrysina, E.D, Kosmopoulou, M.N, Tiraidis, C, Kardakaris, R, Bischler, N, Leonidas, D.D, Hadady, Z, Somsak, L, Docsa, T, Gergely, P, Oikonomakos, N.G. | | Deposit date: | 2004-09-30 | | Release date: | 2005-03-15 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Kinetic and crystallographic studies on 2-(beta-D-glucopyranosyl)-5-methyl-1, 3, 4-oxadiazole, -benzothiazole, and -benzimidazole, inhibitors of muscle glycogen phosphorylase b. Evidence for a new binding site
Protein Sci., 14, 2005
|
|
2CJS
 
 | | Structural Basis for a Munc13-1 Homodimer - Munc13-1 - RIM Heterodimer Switch: C2-domains as Versatile Protein-Protein Interaction Modules | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, REGULATING SYNAPTIC MEMBRANE EXOCYTOSIS PROTEIN 2, ... | | Authors: | Lu, J, Machius, M, Dulubova, I, Dai, H, Sudhof, T.C, Tomchick, D.R, Rizo, J. | | Deposit date: | 2006-04-06 | | Release date: | 2006-06-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural Basis for a Munc13-1 Homodimer to Munc13-1/Rim Heterodimer Switch.
Plos Biol., 4, 2006
|
|
4KBS
 
 | | Crystal structure of human ceramide-1-phosphate transfer protein (CPTP) in complex with 12:0 phosphatidic acid (12:0 PA) | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1,2-ETHANEDIOL, Glycolipid transfer protein domain-containing protein 1 | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-04-23 | | Release date: | 2013-07-17 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Non-vesicular trafficking by a ceramide-1-phosphate transfer protein regulates eicosanoids.
Nature, 500, 2013
|
|
3OHI
 
 | | Structure of Giardia fructose-1,6-biphosphate aldolase in complex with 3-hydroxy-2-pyridone | | Descriptor: | ({3-hydroxy-2-oxo-4-[2-(phosphonooxy)ethyl]pyridin-1(2H)-yl}methyl)phosphonic acid, Putative fructose-1,6-bisphosphate aldolase, ZINC ION | | Authors: | Herzberg, O, Galkin, A. | | Deposit date: | 2010-08-17 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational design, synthesis and evaluation of first generation inhibitors of the Giardia lamblia fructose-1,6-biphosphate aldolase.
J.Inorg.Biochem., 105, 2010
|
|
2R11
 
 | |
3SE9
 
 | | Crystal structure of broadly and potently neutralizing antibody VRC-PG04 in complex with HIV-1 gp120 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kwong, P.D, Zhou, T. | | Deposit date: | 2011-06-10 | | Release date: | 2011-08-10 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing.
Science, 333, 2011
|
|
3CLM
 
 | |
1TXZ
 
 | | Crystal structure of yeast ymx7, an ADP-ribose-1''-monophosphatase, complexed with ADP-ribose | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5-DIPHOSPHORIBOSE, Hypothetical 32.1 kDa protein in ADH3-RCA1 intergenic region, ... | | Authors: | Kumaran, D, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-07-06 | | Release date: | 2004-11-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and mechanism of ADP-ribose-1''-monophosphatase (Appr-1''-pase), a ubiquitous cellular processing enzyme.
Protein Sci., 14, 2005
|
|
