7A8U
 
 | | Crystal structure of sarcomeric protein FATZ-1 (d91-FATZ-1 construct) in complex with rod domain of alpha-actinin-2 | | Descriptor: | Alpha-actinin-2, Myozenin-1 | | Authors: | Sponga, A, Arolas, J.L, Rodriguez Chamorro, A, Mlynek, G, Hollerl, E, Schreiner, C, Pedron, M, Kostan, J, Ribeiro, E.A, Djinovic-Carugo, K. | | Deposit date: | 2020-08-31 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.802 Å) | | Cite: | Order from disorder in the sarcomere: FATZ forms a fuzzy but tight complex and phase-separated condensates with alpha-actinin.
Sci Adv, 7, 2021
|
|
6LTF
 
 | |
6V8Z
 
 | |
6MFI
 
 | | MIM-2 Metallo-Beta-Lactamase | | Descriptor: | Metallo-beta-lactamase, ZINC ION | | Authors: | Selleck, C, Guddat, L, Schenk, G, Monteiro Pedroso, M. | | Deposit date: | 2018-09-11 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.839 Å) | | Cite: | characterization of the B3 MBLs MIM-1 and MIM-2 from environmental microorganisms.
Chemistry, 23, 2017
|
|
6ZWW
 
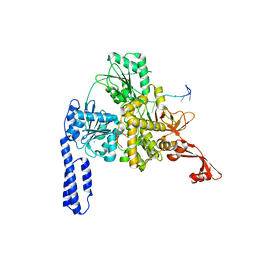 | | Crystal structure of E. coli RNA helicase HrpA in complex with RNA | | Descriptor: | ATP-dependent RNA helicase HrpA, CALCIUM ION, ssRNA | | Authors: | Grass, L.M, Wollenhaupt, J, Barthel, T, Loll, B, Wahl, M.C. | | Deposit date: | 2020-07-29 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Large-scale ratcheting in a bacterial DEAH/RHA-type RNA helicase that modulates antibiotics susceptibility.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MT7
 
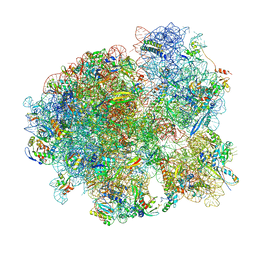 | | Mtb 70S with P and E site tRNAs | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Cui, Z, Zhang, J. | | Deposit date: | 2021-05-13 | | Release date: | 2022-02-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Interplay between an ATP-binding cassette F protein and the ribosome from Mycobacterium tuberculosis.
Nat Commun, 13, 2022
|
|
6V9N
 
 | |
8DNN
 
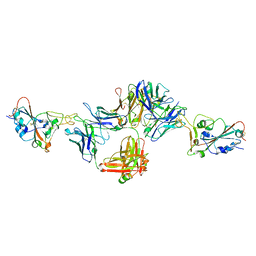 | | Crystal structure of neutralizing antibody 80 in complex with SARS-CoV-2 receptor binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 80 FAB HEAVY CHAIN, 80 FAB LIGHT CHAIN, ... | | Authors: | Muthuraman, K, Kucharska, I, Ivanochko, D, Julien, J.P. | | Deposit date: | 2022-07-11 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | A multi-specific, multi-affinity antibody platform neutralizes sarbecoviruses and confers protection against SARS-CoV-2 in vivo.
Sci Transl Med, 15, 2023
|
|
6P5H
 
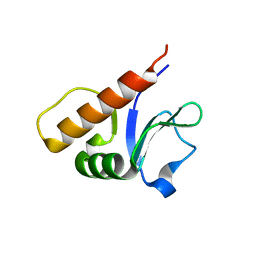 | |
6MFS
 
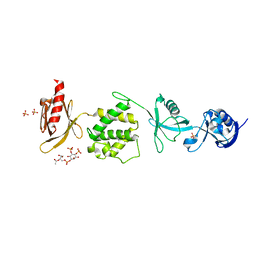 | | Mouse talin1 residues 1-138 fused to residues 169-400 in complex with phosphatidylinositol 4,5-bisphosphate (PIP2) | | Descriptor: | PHOSPHATE ION, Talin-1 fusion, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Izard, T, Chinthalapudi, K, Rangarajan, E.S. | | Deposit date: | 2018-09-12 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The interaction of talin with the cell membrane is essential for integrin activation and focal adhesion formation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6UTN
 
 | | Native E. coli Glyceraldehyde 3-phosphate dehydrogenase | | Descriptor: | ACETATE ION, Glyceraldehyde-3-phosphate dehydrogenase, PHOSPHATE ION, ... | | Authors: | Rodriguez-Hernandez, A, Romo-Arevalo, E, Rodriguez-Romero, A. | | Deposit date: | 2019-10-29 | | Release date: | 2019-12-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A Novel Substrate-Binding Site in the X-Ray Structure of an Oxidized E. coli Glyceraldehyde 3-Phosphate Dehydrogenase Elucidated by Single-Wavelength Anomalous Dispersion
Crystals, 9, 2019
|
|
6P5O
 
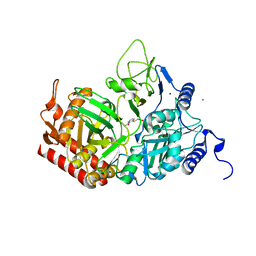 | |
7M26
 
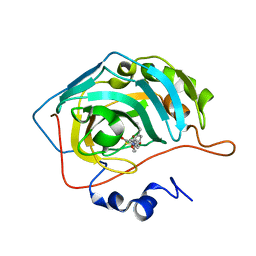 | | Human carbonic anhydrase II in complex with pioglitazone | | Descriptor: | (5R)-5-{4-[2-(5-ethylpyridin-2-yl)ethoxy]benzyl}-1,3-thiazolidine-2,4-dione, Carbonic anhydrase 2, ZINC ION | | Authors: | Mueller, S.L, Peat, T.S. | | Deposit date: | 2021-03-16 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Glitazone Class of Drugs as Carbonic Anhydrase Inhibitors-A Spin-Off Discovery from Fragment Screening.
Molecules, 26, 2021
|
|
6P5R
 
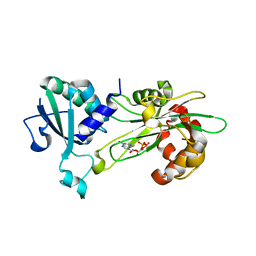 | | Structure of T. brucei MERS1-GDP complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Mitochondrial edited mRNA stability factor 1 | | Authors: | Schumacher, M.A. | | Deposit date: | 2019-05-30 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structures of MERS1, the 5' processing enzyme of mitochondrial mRNAs inTrypanosoma brucei.
Rna, 26, 2020
|
|
6UTS
 
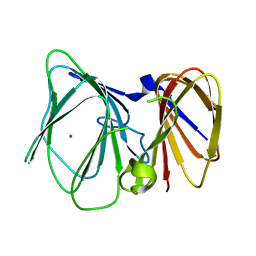 | |
7M24
 
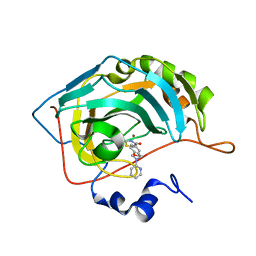 | |
6MFV
 
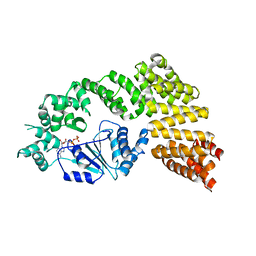 | | Crystal structure of the Signal Transduction ATPase with Numerous Domains (STAND) protein with a tetratricopeptide repeat sensor PH0952 from Pyrococcus horikoshii | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, tetratricopeptide repeat sensor PH0952 | | Authors: | Lisa, M.N, Alzari, P.M, Haouz, A, Danot, O. | | Deposit date: | 2018-09-12 | | Release date: | 2019-02-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Double autoinhibition mechanism of signal transduction ATPases with numerous domains (STAND) with a tetratricopeptide repeat sensor.
Nucleic Acids Res., 47, 2019
|
|
6ZQK
 
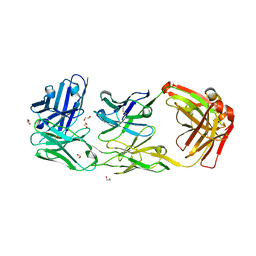 | | HER2-binding scFv-Fab fusion 841 | | Descriptor: | 1,2-ETHANEDIOL, 841 heavy chain, 841 light chain | | Authors: | Kast, F, Schwill, M, Stueber, J.C, Pfundstein, S, Nagy-Davidescu, G, Monne Rodriguez, J.M, Seehusen, F, Richter, C.P, Honegger, A, Hartmann, K.P, Weber, T.G, Kroener, F, Ernst, P, Piehler, J, Plueckthun, A. | | Deposit date: | 2020-07-09 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Engineering an anti-HER2 biparatopic antibody with a multimodal mechanism of action.
Nat Commun, 12, 2021
|
|
6UTV
 
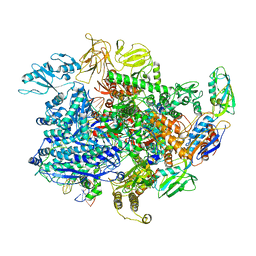 | | E. coli sigma-S transcription initiation complex with a 6-nt RNA ("Fresh" crystal soaked with CTP, UTP, GTP, and ddATP for 150 seconds) | | Descriptor: | DIPHOSPHATE, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Zuo, Y, De, S, Steitz, T.A. | | Deposit date: | 2019-10-30 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Structural Insights into Transcription Initiation from De Novo RNA Synthesis to Transitioning into Elongation.
Iscience, 23, 2020
|
|
8E5E
 
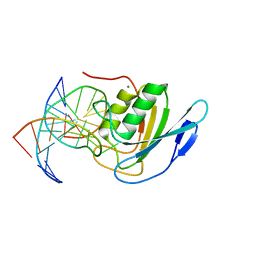 | | Crystal structure of double-stranded DNA deaminase toxin DddA in complex with DNA with the target cytosine flipped into the active site | | Descriptor: | DNA (5'-D(*GP*CP*AP*AP*CP*GP*TP*CP*CP*GP*GP*TP*AP*C)-3'), DNA (5'-D(*GP*TP*AP*CP*CP*GP*GP*AP*CP*GP*TP*TP*GP*C)-3'), Double-stranded DNA deaminase toxin A, ... | | Authors: | Yin, L, Shi, K, Aihara, H. | | Deposit date: | 2022-08-21 | | Release date: | 2023-05-24 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural basis of sequence-specific cytosine deamination by double-stranded DNA deaminase toxin DddA.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6M4O
 
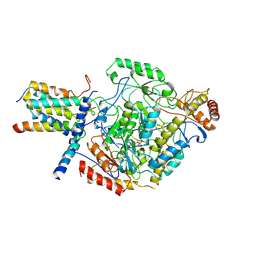 | | Cryo-EM structure of the monomeric SPT-ORMDL3 complex | | Descriptor: | ORM1-like protein 3, PYRIDOXAL-5'-PHOSPHATE, Serine palmitoyltransferase 1, ... | | Authors: | Li, S.S, Xie, T, Wang, L, Gong, X. | | Deposit date: | 2020-03-08 | | Release date: | 2021-02-10 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into the assembly and substrate selectivity of human SPT-ORMDL3 complex.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7MK5
 
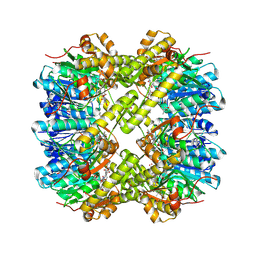 | | Crystal structure of Escherichia coli ClpP covalently inhibited by clipibicyclene | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-[(1E)-3-{[(2E,4E,6E,8S)-8-hydroxy-4-methyldeca-2,4,6-trienoyl]amino}-3-oxoprop-1-en-1-yl]azete-1(2H)-carboxylic acid, ACETATE ION, ... | | Authors: | Culp, E.J, Sychantha, D, Hobson, C, Pawlowski, A.J, Prehna, G, Wright, G.D. | | Deposit date: | 2021-04-21 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | ClpP inhibitors are produced by a widespread family of bacterial gene clusters.
Nat Microbiol, 7, 2022
|
|
6P5V
 
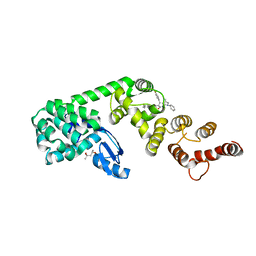 | | Structure of DCN1 bound to N-((4S,5S)-7-ethyl-4-(4-fluorophenyl)-3-methyl-6-oxo-1-phenyl-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl)-3-methylbenzamide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lysozyme,DCN1-like protein 1 fusion, N-[(4S,5S)-1-[(1S)-cyclohex-3-en-1-yl]-7-ethyl-4-(4-fluorophenyl)-3-methyl-6-oxo-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl]-3-methylbenzamide | | Authors: | Guy, R.K, Kim, H.S, Hammill, J.T, Scott, D.C, Schulman, B.A. | | Deposit date: | 2019-05-31 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | Discovery of Novel Pyrazolo-pyridone DCN1 Inhibitors Controlling Cullin Neddylation.
J.Med.Chem., 62, 2019
|
|
8DXL
 
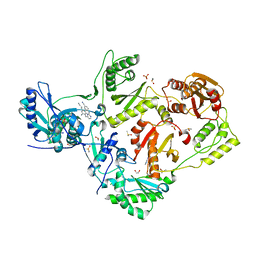 | | HIV-1 reverse transcriptase/rilpivirine with bound fragment 4-iodopyrazole at multiple sites | | Descriptor: | 1,2-ETHANEDIOL, 4-IODOPYRAZOLE, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, ... | | Authors: | Chopra, A, Ruiz, F.X, Bauman, J.D, Arnold, E. | | Deposit date: | 2022-08-02 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Halo Library, a Tool for Rapid Identification of Ligand Binding Sites on Proteins Using Crystallographic Fragment Screening.
J.Med.Chem., 66, 2023
|
|
6V9Z
 
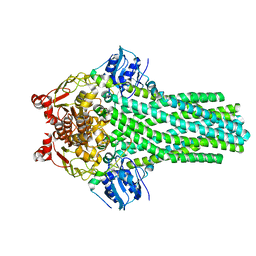 | |
