7RE2
 
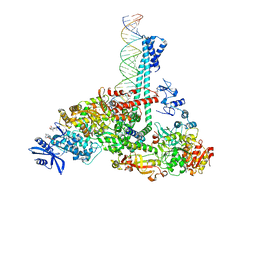 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(1)-RTC | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-12-01 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RDY
 
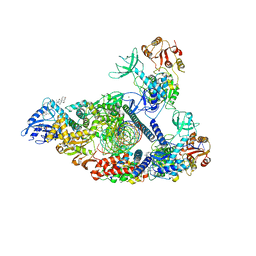 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC - engaged class | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-12-01 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RE0
 
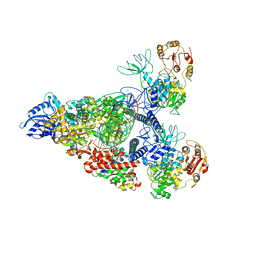 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC - swiveled class | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Helicase, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-12-01 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8GW1
 
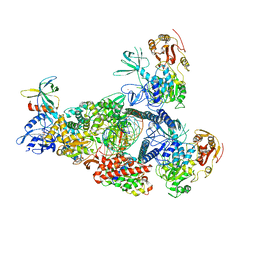 | | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analogue inhibitors | | Descriptor: | Helicase, MANGANESE (II) ION, Non-structural protein 7, ... | | Authors: | Yan, L, Rao, Z, Lou, Z. | | Deposit date: | 2022-09-16 | | Release date: | 2023-10-25 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GY6
 
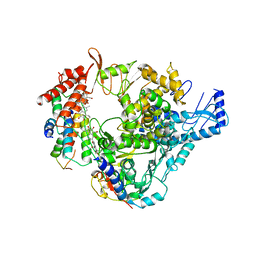 | |
8G6R
 
 | | Porcine epidemic diarrhea virus core polymerase complex | | Descriptor: | RNA (5'-R(P*AP*AP*GP*AP*AP*GP*CP*UP*AP*UP*UP*AP*AP*AP*AP*UP*CP*AP*CP*A)-3'), RNA (5'-R(P*GP*GP*UP*UP*GP*UP*GP*AP*UP*UP*UP*UP*AP*AP*UP*AP*GP*CP*UP*U)-3'), ZINC ION, ... | | Authors: | Anderson, T.K, Kirchdoerfer, R.N. | | Deposit date: | 2023-02-15 | | Release date: | 2023-03-29 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | An alphacoronavirus polymerase structure reveals conserved co-factor functions.
Biorxiv, 2023
|
|
8GWM
 
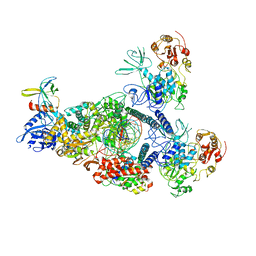 | | SARS-CoV-2 E-RTC bound with MMP-nsp9 and GMPPNP | | Descriptor: | 2'-deoxy-2'-fluoro-2'-methyluridine 5'-(trihydrogen diphosphate), Helicase, Non-structural protein 7, ... | | Authors: | Yan, L.M, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2022-09-17 | | Release date: | 2024-02-28 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
5FJ7
 
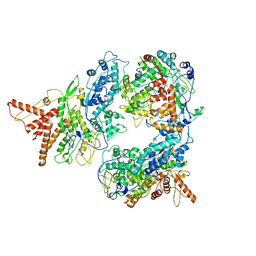 | | Structure of the P2 polymerase inside in vitro assembled bacteriophage phi6 polymerase complex, with P1 included | | Descriptor: | MAJOR INNER PROTEIN P1, MANGANESE (II) ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Ilca, S, Kotecha, A, Sun, X, Poranen, M.P, Stuart, D.I, Huiskonen, J.T. | | Deposit date: | 2015-10-06 | | Release date: | 2015-11-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Localized Reconstruction of Subunits from Electron Cryomicroscopy Images of Macromolecular Complexes.
Nat.Commun., 6, 2015
|
|
7BV2
 
 | | The nsp12-nsp7-nsp8 complex bound to the template-primer RNA and triphosphate form of Remdesivir(RTP) | | Descriptor: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yin, W, Mao, C, Luan, X, Shen, D, Shen, Q, Su, H, Wang, X, Zhou, F, Zhao, W, Gao, M, Chang, S, Xie, Y.C, Tian, G, Jiang, H.W, Tao, S.C, Shen, J, Jiang, Y, Jiang, H, Xu, Y, Zhang, S, Zhang, Y, Xu, H.E. | | Deposit date: | 2020-04-09 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir.
Science, 368, 2020
|
|
7BV1
 
 | | Cryo-EM structure of the apo nsp12-nsp7-nsp8 complex | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA-directed RNA polymerase, ... | | Authors: | Yin, W, Mao, C, Luan, X, Shen, D, Shen, Q, Su, H, Wang, X, Zhou, F, Zhao, W, Gao, M, Chang, S, Xie, Y.C, Tian, G, Jiang, H.W, Tao, S.C, Shen, J, Jiang, Y, Jiang, H, Xu, Y, Zhang, S, Zhang, Y, Xu, H.E. | | Deposit date: | 2020-04-09 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir.
Science, 368, 2020
|
|
7BZF
 
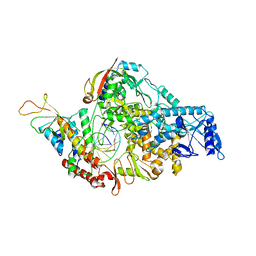 | | COVID-19 RNA-dependent RNA polymerase post-translocated catalytic complex | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA (31-MER), ... | | Authors: | Wang, Q, Gao, Y, Ji, W, Mu, A, Rao, Z. | | Deposit date: | 2020-04-27 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural Basis for RNA Replication by the SARS-CoV-2 Polymerase.
Cell, 182, 2020
|
|
7CXN
 
 | | Architecture of a SARS-CoV-2 mini replication and transcription complex | | Descriptor: | Helicase, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yan, L, Zhang, Y, Ge, J, Zheng, L, Gao, Y, Wang, T, Jia, Z, Wang, H, Huang, Y, Li, M, Wang, Q, Rao, Z, Lou, Z. | | Deposit date: | 2020-09-02 | | Release date: | 2020-11-04 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Architecture of a SARS-CoV-2 mini replication and transcription complex.
Nat Commun, 11, 2020
|
|
7CXM
 
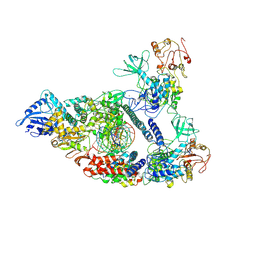 | | Architecture of a SARS-CoV-2 mini replication and transcription complex | | Descriptor: | Helicase, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yan, L, Zhang, Y, Ge, J, Zheng, L, Gao, Y, Wang, T, Jia, Z, Wang, H, Huang, Y, Li, M, Wang, Q, Rao, Z, Lou, Z. | | Deposit date: | 2020-09-02 | | Release date: | 2020-11-04 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Architecture of a SARS-CoV-2 mini replication and transcription complex.
Nat Commun, 11, 2020
|
|
7DTE
 
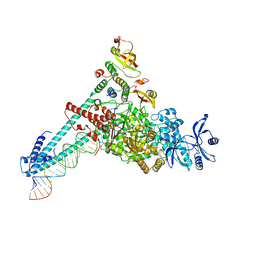 | | SARS-CoV-2 RdRP catalytic complex with T33-1 RNA | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA (33-MER), ... | | Authors: | Wang, Q, Gong, P. | | Deposit date: | 2021-01-04 | | Release date: | 2021-10-20 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Remdesivir overcomes the S861 roadblock in SARS-CoV-2 polymerase elongation complex.
Cell Rep, 37, 2021
|
|
7DFH
 
 | |
7CYQ
 
 | | Cryo-EM structure of an extended SARS-CoV-2 replication and transcription complex reveals an intermediate state in cap synthesis | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Helicase, MAGNESIUM ION, ... | | Authors: | Yan, L, Ge, J, Zheng, L, Zhang, Y, Gao, Y, Wang, T, Wang, H, Huang, Y, Li, M, Wang, Q, Rao, Z, Lou, Z. | | Deposit date: | 2020-09-04 | | Release date: | 2020-12-09 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cryo-EM Structure of an Extended SARS-CoV-2 Replication and Transcription Complex Reveals an Intermediate State in Cap Synthesis.
Cell, 184, 2021
|
|
7C2K
 
 | | COVID-19 RNA-dependent RNA polymerase pre-translocated catalytic complex | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA (29-MER), ... | | Authors: | Wang, Q, Gao, Y, Ji, W, Mu, A, Rao, Z. | | Deposit date: | 2020-05-07 | | Release date: | 2020-06-03 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural Basis for RNA Replication by the SARS-CoV-2 Polymerase.
Cell, 182, 2020
|
|
7BTF
 
 | | SARS-CoV-2 RNA-dependent RNA polymerase in complex with cofactors in reduced condition | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA-directed RNA polymerase, ... | | Authors: | Gao, Y, Yan, L, Huang, Y, Liu, F, Cao, L, Wang, T, Wang, Q, Lou, Z, Rao, Z. | | Deposit date: | 2020-04-01 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structure of the RNA-dependent RNA polymerase from COVID-19 virus.
Science, 368, 2020
|
|
7ED5
 
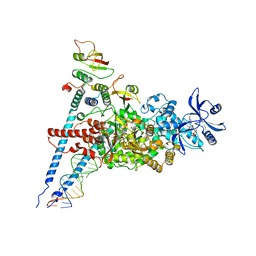 | | A dual mechanism of action of AT-527 against SARS-CoV-2 polymerase | | Descriptor: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Shannon, A, Fattorini, V, Sama, B, Selisko, B, Feracci, M, Falcou, C, Gauffre, P, El Kazzi, P, Delpal, A, Decroly, E, Alvarez, K, Eydoux, C, Guillemot, J.-C, Moussa, A, Good, S, Colla, P, Lin, K, Sommadossi, J.-P, Zhu, Y.X, Yan, X.D, Shi, H, Ferron, F, Canard, B. | | Deposit date: | 2021-03-15 | | Release date: | 2022-02-16 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | A dual mechanism of action of AT-527 against SARS-CoV-2 polymerase.
Nat Commun, 13, 2022
|
|
7EIZ
 
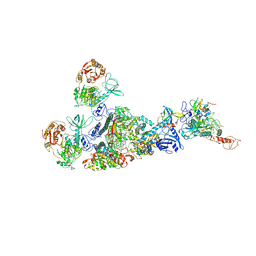 | | Coupling of N7-methyltransferase and 3'-5' exoribonuclease with SARS-CoV-2 polymerase reveals mechanisms for capping and proofreading | | Descriptor: | Helicase, MAGNESIUM ION, Non-structural protein 10, ... | | Authors: | Yan, L, Yang, Y.X, Li, M.Y, Zhang, Y, Zheng, L.T, Ge, J, Huang, Y.C, Liu, Z.Y, Wang, T, Gao, S, Zhang, R, Huang, Y.Y, Guddat, L.W, Gao, Y, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2021-04-01 | | Release date: | 2021-09-22 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Coupling of N7-methyltransferase and 3'-5' exoribonuclease with SARS-CoV-2 polymerase reveals mechanisms for capping and proofreading
Cell, 184, 2021
|
|
2IJD
 
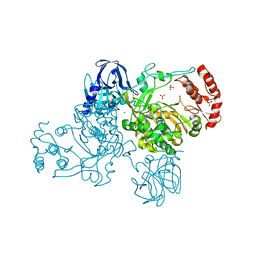 | | Crystal Structure of the Poliovirus Precursor Protein 3CD | | Descriptor: | Picornain 3C, RNA-directed RNA polymerase, SULFATE ION, ... | | Authors: | Marcotte, L.L, Gohara, D.W, Filman, D.J, Hogle, J.M. | | Deposit date: | 2006-09-29 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of poliovirus 3CD: virally-encoded protease and precursor to the RNA-dependent RNA polymerase.
J.Virol., 81, 2007
|
|
7O80
 
 | | Rabbit 80S ribosome in complex with eRF1 and ABCE1 stalled at the STOP codon in the mutated SARS-CoV-2 slippery site | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Bhatt, P.R, Scaiola, A, Leibundgut, M.A, Atkins, J.F, Ban, N. | | Deposit date: | 2021-04-14 | | Release date: | 2021-06-02 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome.
Science, 372, 2021
|
|
7O81
 
 | | Rabbit 80S ribosome colliding in another ribosome stalled by the SARS-CoV-2 pseudoknot | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Bhatt, P.R, Scaiola, A, Leibundgut, M.A, Atkins, J.F, Ban, N. | | Deposit date: | 2021-04-14 | | Release date: | 2021-06-02 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome.
Science, 372, 2021
|
|
7O7Z
 
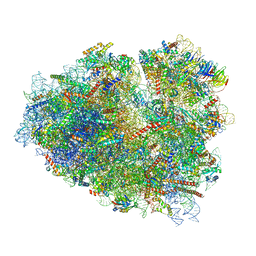 | | Rabbit 80S ribosome stalled close to the mutated SARS-CoV-2 slippery site by a pseudoknot (classified for pseudoknot) | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Bhatt, P.R, Scaiola, A, Leibundgut, M.A, Atkins, J.F, Ban, N. | | Deposit date: | 2021-04-14 | | Release date: | 2021-06-02 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome.
Science, 372, 2021
|
|
7O7Y
 
 | | Rabbit 80S ribosome stalled close to the mutated SARS-CoV-2 slippery site by a pseudoknot (high resolution) | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Bhatt, P.R, Scaiola, A, Leibundgut, M.A, Atkins, J.F, Ban, N. | | Deposit date: | 2021-04-14 | | Release date: | 2021-06-02 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome.
Science, 372, 2021
|
|
