7DS7
 
 | | The Crystal Structure of Leaf-branch compost cutinase from Biortus. | | Descriptor: | CITRIC ACID, GLYCEROL, IMIDAZOLE, ... | | Authors: | Wang, F, Lv, Z, Cheng, W, Lin, D, Chu, F, Xu, X, Tan, J. | | Deposit date: | 2020-12-30 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Crystal Structure of Leaf-branch compost cutinase from Biortus.
To Be Published
|
|
1ULN
 
 | | Crystal Structure of Pokeweed Lectin-D1 | | Descriptor: | lectin-D | | Authors: | Fujii, T, Hayashida, M, Hamasu, M, Ishiguro, M, Hata, Y. | | Deposit date: | 2003-09-16 | | Release date: | 2004-04-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of two lectins from the roots of pokeweed (Phytolacca americana).
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1ULM
 
 | | Crystal Structure of Pokeweed Lectin-D2 complexed with tri-N-acetylchitotriose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, lectin-D2 | | Authors: | Hayashida, M, Fujii, T, Ishiguro, M, Hata, Y. | | Deposit date: | 2003-09-12 | | Release date: | 2003-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Similarity between protein-protein and protein-carbohydrate interactions, revealed by two crystal structures of lectins from the roots of pokeweed.
J.Mol.Biol., 334, 2003
|
|
7D7S
 
 | |
7DA5
 
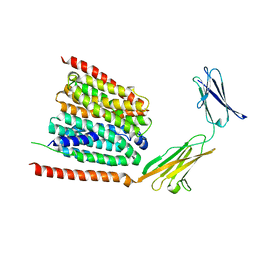 | | Cryo-EM structure of the human MCT1 D309N mutant in complex with Basigin-2 in the inward-open conformation. | | Descriptor: | Basigin, Monocarboxylate transporter 1 | | Authors: | Wang, N, Jiang, X, Zhang, S, Zhu, A, Yuan, Y, Lei, J, Yan, C. | | Deposit date: | 2020-10-14 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of human monocarboxylate transporter 1 inhibition by anti-cancer drug candidates.
Cell, 184, 2021
|
|
1TOJ
 
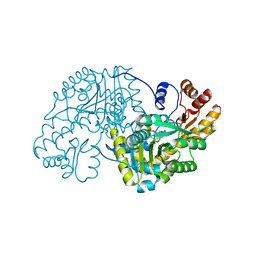 | | Hydrocinnamic acid-bound structure of SRHEPT mutant of E. coli aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, HYDROCINNAMIC ACID | | Authors: | Chow, M.A, McElroy, K.E, Corbett, K.D, Berger, J.M, Kirsch, J.F. | | Deposit date: | 2004-06-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Narrowing substrate specificity in a directly evolved enzyme: the A293D mutant of aspartate aminotransferase
Biochemistry, 43, 2004
|
|
1TN6
 
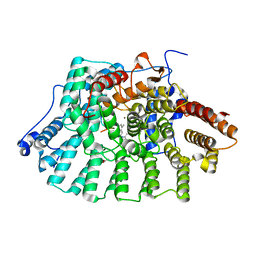 | | Protein Farnesyltransferase Complexed with a Rap2a Peptide Substrate and a FPP Analog at 1.8A Resolution | | Descriptor: | ACETIC ACID, Protein farnesyltransferase alpha subunit, Protein farnesyltransferase beta subunit, ... | | Authors: | Reid, T.S, Terry, K.L, Casey, P.J, Beese, L.S. | | Deposit date: | 2004-06-11 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic analysis of CaaX prenyltransferases complexed with substrates defines rules of protein substrate selectivity
J.Mol.Biol., 343, 2004
|
|
1TO2
 
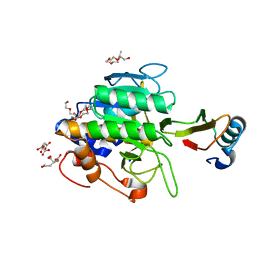 | | crystal structure of the complex of subtilisin BPN' with chymotrypsin inhibitor 2 M59K, in pH 9 cryosoak | | Descriptor: | CALCIUM ION, CITRIC ACID, POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Radisky, E.S, Kwan, G, Karen Lu, C.J, Koshland Jr, D.E. | | Deposit date: | 2004-06-11 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Binding, Proteolytic, and Crystallographic Analyses of Mutations at the Protease-Inhibitor Interface of the Subtilisin BPN'/Chymotrypsin Inhibitor 2 Complex(,).
Biochemistry, 43, 2004
|
|
7D93
 
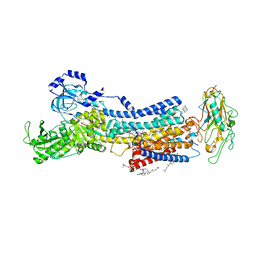 | | Crystal Structure of the Na+,K+-ATPase in the E2P state with bound Mg2+ and anthroylouabain (P2(1)2(1)2(1) symmetry) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kanai, R, Cornelius, F, Ogawa, H, Motoyama, K, Vilsen, B, Toyoshima, C. | | Deposit date: | 2020-10-12 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Binding of cardiotonic steroids to Na + ,K + -ATPase in the E2P state.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7DDI
 
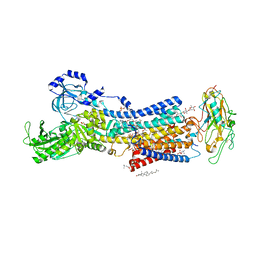 | | Crystal structures of Na+,K+-ATPase in complex with digitoxin | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ogawa, H, Cornelius, F, Kanai, R, Motoyama, K, Vilsen, B, Toyoshima, C. | | Deposit date: | 2020-10-29 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.72 Å) | | Cite: | Binding of cardiotonic steroids to Na + ,K + -ATPase in the E2P state.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1TPY
 
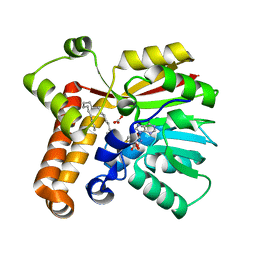 | |
7D90
 
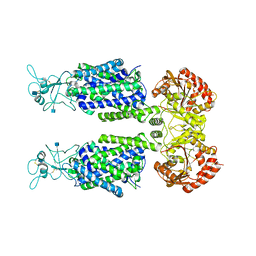 | | human potassium-chloride co-transporter KCC3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, potassium-chloride cotransporter 3 | | Authors: | Xie, Y, Chang, S, Zhao, C, Ye, S, Guo, J. | | Deposit date: | 2020-10-12 | | Release date: | 2020-12-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures and an activation mechanism of human potassium-chloride cotransporters.
Sci Adv, 6, 2020
|
|
1TN0
 
 | | Structure of bacterorhodopsin mutant A51P | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Yohannan, S, Yang, D, Faham, S, Boulting, G, Whitelegge, J, Bowie, J.U. | | Deposit date: | 2004-06-11 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Proline substitutions are not easily accommodated in a membrane protein
J.Mol.Biol., 341, 2004
|
|
7DDK
 
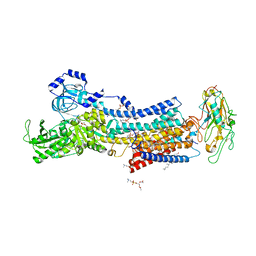 | | Crystal structures of Na+,K+-ATPase in complex with rostafuroxin | | Descriptor: | (3S,5R,8R,9S,10S,13S,14S,17S)-17-(furan-3-yl)-10,13-dimethyl-2,3,4,5,6,7,8,9,11,12,15,16-dodecahydro-1H-cyclopenta[a]phenanthrene-3,14,17-triol, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ogawa, H, Cornelius, F, Kanai, R, Motoyama, K, Vilsen, B, Toyoshima, C. | | Deposit date: | 2020-10-29 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Binding of cardiotonic steroids to Na + ,K + -ATPase in the E2P state.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7DFP
 
 | | Human dopamine D2 receptor in complex with spiperone | | Descriptor: | 8-[4-(4-fluorophenyl)-4-oxidanylidene-butyl]-1-phenyl-1,3,8-triazaspiro[4.5]decan-4-one, D(2) dopamine receptor,Soluble cytochrome b562, FabH, ... | | Authors: | Im, D, Shimamura, T, Iwata, S. | | Deposit date: | 2020-11-09 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the dopamine D 2 receptor in complex with the antipsychotic drug spiperone.
Nat Commun, 11, 2020
|
|
7DI8
 
 | |
7D8Z
 
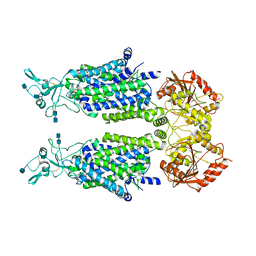 | | human potassium-chloride co-transporter KCC2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, potassium-chloride cotransporter 2 | | Authors: | Xie, Y, Chang, S, Zhao, C, Ye, S, Guo, J. | | Deposit date: | 2020-10-11 | | Release date: | 2020-12-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures and an activation mechanism of human potassium-chloride cotransporters.
Sci Adv, 6, 2020
|
|
7DMQ
 
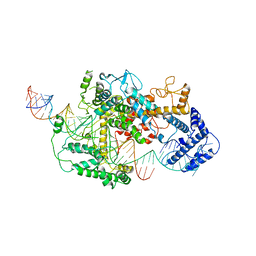 | | Cryo-EM structure of LshCas13a-crRNA-anti-tag RNA complex | | Descriptor: | Anti-tag target RNA, CRISPR RNA, CRISPR/Cas system Cas13a | | Authors: | Wang, B, Zhang, T, Ding, J, Patel, D.J, Yang, H. | | Deposit date: | 2020-12-05 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis for self-cleavage prevention by tag:anti-tag pairing complementarity in type VI Cas13 CRISPR systems.
Mol.Cell, 81, 2021
|
|
1TOI
 
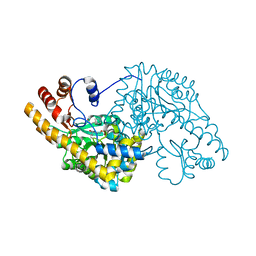 | | Hydrocinnamic acid-bound structure of Hexamutant + A293D mutant of E. coli aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, HYDROCINNAMIC ACID | | Authors: | Chow, M.A, McElroy, K.E, Corbett, K.D, Berger, J.M, Kirsch, J.F. | | Deposit date: | 2004-06-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Narrowing substrate specificity in a directly evolved enzyme: the A293D mutant of aspartate aminotransferase
Biochemistry, 43, 2004
|
|
7D58
 
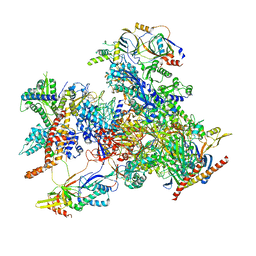 | | cryo-EM structure of human RNA polymerase III in elongating state | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Wang, Q, Wan, F, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2020-09-25 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into transcriptional regulation of human RNA polymerase III.
Nat.Struct.Mol.Biol., 28, 2021
|
|
1TN8
 
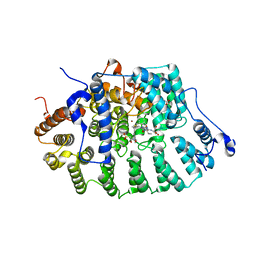 | | Protein Farnesyltransferase Complexed with a H-Ras Peptide Substrate and a FPP Analog at 2.25A Resolution | | Descriptor: | ACETIC ACID, Protein farnesyltransferase beta subunit, Protein farnesyltransferase/geranylgeranyltransferase type I alpha subunit, ... | | Authors: | Reid, T.S, Terry, K.L, Casey, P.J, Beese, L.S. | | Deposit date: | 2004-06-11 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystallographic analysis of CaaX prenyltransferases complexed with substrates defines rules of protein substrate selectivity.
J.Mol.Biol., 343, 2004
|
|
7D59
 
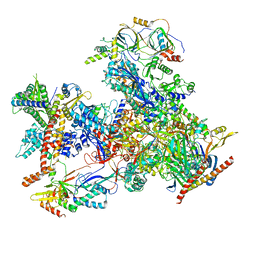 | | cryo-EM structure of human RNA polymerase III in apo state | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Wang, Q, Wan, F, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2020-09-25 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into transcriptional regulation of human RNA polymerase III.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7EPA
 
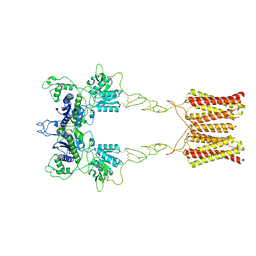 | | Cryo-EM structure of inactive mGlu2 homodimer | | Descriptor: | Metabotropic glutamate receptor 2 | | Authors: | Du, J, Wang, D, Fan, H, Tai, L, Lin, S, Han, S, Sun, F, Wu, B, Zhao, Q. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of human mGlu2 and mGlu7 homo- and heterodimers.
Nature, 594, 2021
|
|
7ETW
 
 | | Cryo-EM structure of Scap/Insig complex in the present of digitonin. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Digitonin, Insulin-induced gene 2 protein, ... | | Authors: | Yan, R, Cao, P, Song, W, Li, Y, Wang, T, Qian, H, Yan, C, Yan, N. | | Deposit date: | 2021-05-14 | | Release date: | 2021-06-23 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for sterol sensing by Scap and Insig
Cell Rep, 35, 2021
|
|
7EZZ
 
 | | Crystal structure of Salmonella typhi outer membrane phospholipase (OMPLA) dimer with bound calcium | | Descriptor: | CALCIUM ION, DODECANE, Phospholipase A1, ... | | Authors: | Perumal, P, Raina, R, Sreeshma, N.S, Arockiasamy, A, Sundarabaalaji, N. | | Deposit date: | 2021-06-02 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Crystal structure of Salmonella typhi outer membrane phospholipase (OMPLA) dimer with bound calcium
To Be Published
|
|
