7U24
 
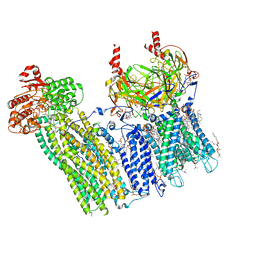 | | Cryo-EM structure of the pancreatic ATP-sensitive potassium channel bound to ATP and glibenclamide with Kir6.2-CTD in the up conformation | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide, ... | | Authors: | Shyng, S.L, Sung, M.W, Driggers, C.M. | | Deposit date: | 2022-02-23 | | Release date: | 2022-08-31 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Ligand-mediated Structural Dynamics of a Mammalian Pancreatic K ATP Channel.
J.Mol.Biol., 434, 2022
|
|
7U6Y
 
 | |
7U7M
 
 | |
2NXX
 
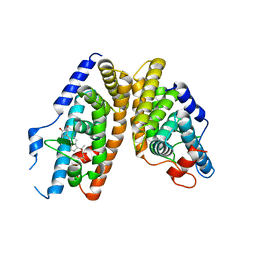 | | Crystal Structure of the Ligand-Binding Domains of the T.castaneum (Coleoptera) Heterodimer EcrUSP Bound to Ponasterone A | | Descriptor: | 2,3,14,20,22-PENTAHYDROXYCHOLEST-7-EN-6-ONE, Ecdysone Receptor (EcR, NRH1), ... | | Authors: | Iwema, T, Billas, I, Moras, D. | | Deposit date: | 2006-11-20 | | Release date: | 2007-10-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural and functional characterization of a novel type of ligand-independent RXR-USP receptor.
Embo J., 26, 2007
|
|
4S15
 
 | | Crystal structure of the orphan nuclear receptor RORalpha ligand-binding domain in complex with 4alpha-caboxyl, 4beta-methyl-zymosterol (4ACD8) | | Descriptor: | (3beta,4alpha,5beta,14beta)-3-hydroxy-4-methylcholesta-8,24-diene-4-carboxylic acid, GLYCEROL, Nuclear receptor ROR-alpha, ... | | Authors: | Huang, P, Santori, F.R, Littman, D.R, Rastinejad, F. | | Deposit date: | 2015-01-08 | | Release date: | 2015-02-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | Identification of Natural ROR gamma Ligands that Regulate the Development of Lymphoid Cells.
Cell Metab, 21, 2015
|
|
226L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | Deposit date: | 1997-06-25 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
3W0I
 
 | | Crystal Structure of Rat VDR Ligand Binding Domain in Complex with Novel Nonsecosteroidal Ligands | | Descriptor: | (2S)-3-{4-[3-(4-{[(2R)-2-hydroxy-3,3-dimethylbutyl]oxy}phenyl)pentan-3-yl]phenoxy}propane-1,2-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 Receptor | | Authors: | Shimizu, T, Asano, L, Kuwabara, N, Ito, I, Waku, T, Yanagisawa, J, Miyachi, H. | | Deposit date: | 2012-10-30 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for vitamin D receptor agonism by novel non-secosteroidal ligands.
Febs Lett., 587, 2013
|
|
8TVZ
 
 | |
1KEO
 
 | | TWISTS AND TURNS OF THE CD-MPR: LIGAND-BOUND VERSUS LIGAND-FREE RECEPTOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, cation-dependent mannose-6-phosphate receptor | | Authors: | Olson, L.J, Zhang, J, Dahms, N.M, Kim, J.J. | | Deposit date: | 2001-11-16 | | Release date: | 2002-01-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Twists and turns of the cation-dependent mannose 6-phosphate receptor. Ligand-bound versus ligand-free receptor
J.Biol.Chem., 277, 2002
|
|
4AP4
 
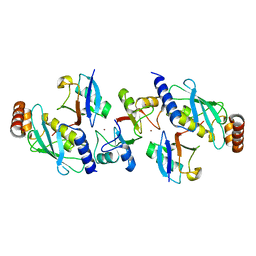 | | Rnf4 - ubch5a - ubiquitin heterotrimeric complex | | Descriptor: | E3 UBIQUITIN LIGASE RNF4, UBIQUITIN C, UBIQUITIN-CONJUGATING ENZYME E2 D1, ... | | Authors: | Plechanovova, A, Hay, R.T, Tatham, M.H, Jaffray, E, Naismith, J.H. | | Deposit date: | 2012-03-30 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure of a Ring E3 Ligase and Ubiquitin-Loaded E2 Primed for Catalysis
Nature, 489, 2012
|
|
7OKQ
 
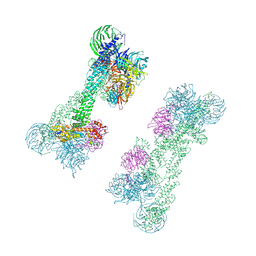 | | Cryo-EM Structure of the DDB1-DCAF1-CUL4A-RBX1 Complex | | Descriptor: | Cullin-4A, DDB1- and CUL4-associated factor 1, DNA damage-binding protein 1, ... | | Authors: | Mohamed, W.I, Schenk, A.D, Kempf, G, Cavadini, S, Thoma, N.H. | | Deposit date: | 2021-05-18 | | Release date: | 2021-10-13 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | The CRL4 DCAF1 cullin-RING ubiquitin ligase is activated following a switch in oligomerization state.
Embo J., 40, 2021
|
|
5ILC
 
 | | The X-ray structure of the adduct formed in the reaction between hen egg white lysozyme a compound 2, a platin(II) compound containing a O, S bidentate ligand | | Descriptor: | ACETATE ION, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Merlino, A, Ferraro, G. | | Deposit date: | 2016-03-04 | | Release date: | 2016-12-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Platinum(ii) O,S complexes as potential metallodrugs against Cisplatin resistance.
Dalton Trans, 45, 2016
|
|
5II3
 
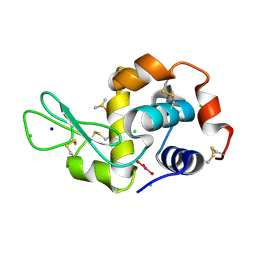 | | The X-ray structure of the adduct formed in the reaction between hen egg white lysozyme and compound 3, a platin(II) compound containing a O, S bidentate ligand | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Lysozyme C, ... | | Authors: | Ferraro, G, Merlino, A. | | Deposit date: | 2016-03-01 | | Release date: | 2016-12-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Platinum(ii) O,S complexes as potential metallodrugs against Cisplatin resistance.
Dalton Trans, 45, 2016
|
|
7B5R
 
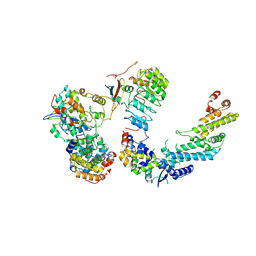 | | Ubiquitin ligation to F-box protein substrates by SCF-RBR E3-E3 super-assembly: CUL1-RBX1-SKP1-SKP2-CKSHS1-Cyclin A-CDK2-p27 | | Descriptor: | Cullin-1, Cyclin-A2, Cyclin-dependent kinase 2, ... | | Authors: | Horn-Ghetko, D, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2020-12-07 | | Release date: | 2021-02-10 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Ubiquitin ligation to F-box protein targets by SCF-RBR E3-E3 super-assembly.
Nature, 590, 2021
|
|
5ILF
 
 | | The X-ray structure of the adduct formed in the reaction between hen egg white lysozyme and compound 4, a platin(II) compound containing a O, S bidentate ligand | | Descriptor: | ACETATE ION, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Merlino, A, Ferraro, G. | | Deposit date: | 2016-03-04 | | Release date: | 2016-12-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Platinum(ii) O,S complexes as potential metallodrugs against Cisplatin resistance.
Dalton Trans, 45, 2016
|
|
1IUH
 
 | | Crystal structure of TT0787 of thermus thermophilus HB8 | | Descriptor: | 2'-5' RNA Ligase | | Authors: | Kato, M, Sakai, H, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-05 | | Release date: | 2003-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the 2'-5' RNA Ligase from Thermus thermophilus HB8
J.MOL.BIOL., 329, 2003
|
|
9DNT
 
 | | Cryo-EM structure of Tom1 (S. cerevisiae) | | Descriptor: | E3 ubiquitin-protein ligase TOM1 | | Authors: | Warner, K.M, Hunkeler, M, Baek, K, Roy Burman, S.S, Fischer, E.S. | | Deposit date: | 2024-09-18 | | Release date: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural ubiquitin contributes to K48 linkage specificity of the HECT ligase Tom1.
Cell Rep, 44, 2025
|
|
7P0K
 
 | | Crystal structure of Autotaxin (ENPP2) with 18F-labeled positron emission tomography ligand | | Descriptor: | 2-[[2-ethyl-6-[4-[2-[(3~{R})-3-fluoranylpyrrolidin-1-yl]-2-oxidanylidene-ethyl]piperazin-1-yl]imidazo[1,2-a]pyridin-3-yl]-methyl-amino]-4-(4-fluorophenyl)-2,3-dihydro-1,3-thiazole-5-carbonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Salgado-Polo, F, Shao, T, Xiao, Z, Van, R, Chen, J, Rong, J, Haider, A, Shao, Y, Josephson, L, Perrakis, A, Liang, S.H. | | Deposit date: | 2021-06-29 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Imaging Autotaxin In Vivo with 18 F-Labeled Positron Emission Tomography Ligands
J Med Chem, 64, 2021
|
|
5IHG
 
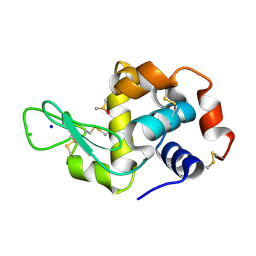 | | The X-ray structure of the adduct formed in the reaction between hen egg white lysozyme a compound I, a platin(II) compound containing a O, S bidentate ligand | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Lysozyme C, ... | | Authors: | Ferraro, G, Merlino, A. | | Deposit date: | 2016-02-29 | | Release date: | 2016-12-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Platinum(ii) O,S complexes as potential metallodrugs against Cisplatin resistance.
Dalton Trans, 45, 2016
|
|
9DRY
 
 | | CHIP U-box dimer bound to Fab 2F1 | | Descriptor: | 2F1 Fab heavy chain, 2F1 Fab light chain, E3 ubiquitin-protein ligase CHIP | | Authors: | Unnikrishnan, A, Southworth, D. | | Deposit date: | 2024-09-26 | | Release date: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (7.02 Å) | | Cite: | Recombinant antibodies inhibit enzymatic activity of the E3 ubiquitin ligase CHIP via multiple mechanisms.
J.Biol.Chem., 301, 2025
|
|
8FH2
 
 | |
8FGZ
 
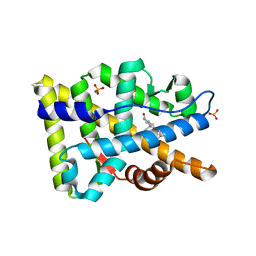 | |
8FGY
 
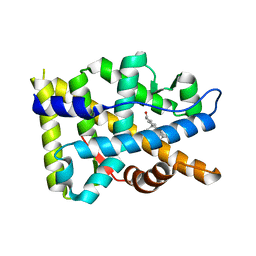 | |
8FH1
 
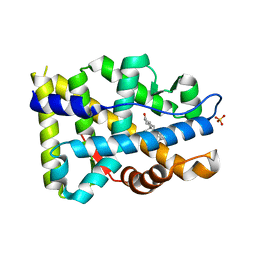 | |
8FH0
 
 | |
