7UG9
 
 | | Crystal structure of RNase AM PHP domain | | Descriptor: | 5'-3' exoribonuclease, MANGANESE (II) ION, SULFATE ION | | Authors: | Doamekpor, S.K, Tong, L. | | Deposit date: | 2022-03-24 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Identification of a novel deFADding activity in human, yeast and bacterial 5' to 3' exoribonucleases.
Nucleic Acids Res., 50, 2022
|
|
6WUG
 
 | |
6WUK
 
 | |
5FG1
 
 | |
5FG0
 
 | |
5ULI
 
 | | Crystal Structure of mouse DXO in complex with (3'-NADP)+ and calcium ion | | Descriptor: | CALCIUM ION, Decapping and exoribonuclease protein, [[(2R,3S,4R,5R)-5-(3-aminocarbonyl-4H-pyridin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Doamekpor, S.K, Tong, L. | | Deposit date: | 2017-01-24 | | Release date: | 2017-05-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 5' End Nicotinamide Adenine Dinucleotide Cap in Human Cells Promotes RNA Decay through DXO-Mediated deNADding.
Cell, 168, 2017
|
|
5ULJ
 
 | |
6WUF
 
 | | Crystal structure of mouse DXO in complex with 3'-FADP | | Descriptor: | 1,2-ETHANEDIOL, Decapping and exoribonuclease protein, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]methyl (2R,3S,4S)-5-(7,8-dimethyl-2,4-dioxo-3,4-dihydrobenzo[g]pteridin-10(2H)-yl)-2,3,4-trihydroxypentyl dihydrogen diphosphate (non-preferred name) | | Authors: | Doamekpor, S.K, Tong, L. | | Deposit date: | 2020-05-04 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | DXO/Rai1 enzymes remove 5'-end FAD and dephospho-CoA caps on RNAs.
Nucleic Acids Res., 48, 2020
|
|
6WRE
 
 | |
6WUI
 
 | | Crystal Structure of mutant S. pombe Rai1 (E150S/E199Q/E239Q) in complex with 3'-FADP | | Descriptor: | Decapping nuclease din1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]methyl (2R,3S,4S)-5-(7,8-dimethyl-2,4-dioxo-3,4-dihydrobenzo[g]pteridin-10(2H)-yl)-2,3,4-trihydroxypentyl dihydrogen diphosphate (non-preferred name) | | Authors: | Doamekpor, S.K, Tong, L. | | Deposit date: | 2020-05-04 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | DXO/Rai1 enzymes remove 5'-end FAD and dephospho-CoA caps on RNAs.
Nucleic Acids Res., 48, 2020
|
|
8FH2
 
 | |
8FGZ
 
 | |
8FH1
 
 | |
8FGY
 
 | |
8FH0
 
 | |
4PZ8
 
 | | PCE1 guanylyltransferase bound to SPT5 CTD | | Descriptor: | SULFATE ION, Transcription elongation factor spt5, mRNA-capping enzyme subunit alpha | | Authors: | Doamekpor, S.K, Lima, C.D. | | Deposit date: | 2014-03-28 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | How an mRNA capping enzyme reads distinct RNA polymerase II and Spt5 CTD phosphorylation codes.
Genes Dev., 28, 2014
|
|
4PZ6
 
 | |
4PZ7
 
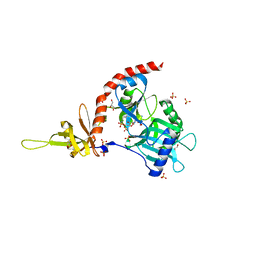 | | PCE1 guanylyltransferase | | Descriptor: | GLYCEROL, SULFATE ION, mRNA-capping enzyme subunit alpha | | Authors: | Doamekpor, S.K, Lima, C.D. | | Deposit date: | 2014-03-28 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.109 Å) | | Cite: | How an mRNA capping enzyme reads distinct RNA polymerase II and Spt5 CTD phosphorylation codes.
Genes Dev., 28, 2014
|
|
4PN0
 
 | |
4PN1
 
 | |
