8ETM
 
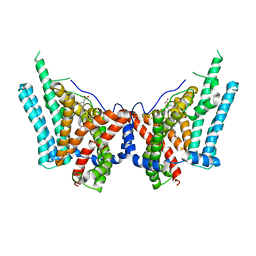 | | Human triacylglycerol synthesizing enzyme DGAT1 in complex with DGAT1IN1 inhibitor | | Descriptor: | Diacylglycerol O-acyltransferase 1, [(1S,4r)-4-{4-[(4S)-2-({[4-(trifluoromethoxy)phenyl]methyl}carbamoyl)imidazo[1,2-a]pyridin-6-yl]phenyl}cyclohexyl]acetic acid | | Authors: | Sui, X, Kun, W, Walther, T, Farese, R, Liao, M. | | Deposit date: | 2022-10-17 | | Release date: | 2023-06-07 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of action for small-molecule inhibitors of triacylglycerol synthesis.
Nat Commun, 14, 2023
|
|
4X11
 
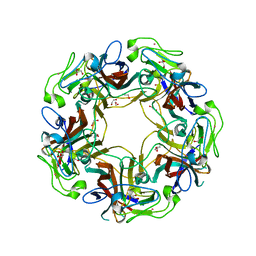 | | JC Polyomavirus genotype 3 VP1 in complex with GD1a oligosaccharide | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Major capsid protein, ... | | Authors: | Stroh, L.J, Stehle, T. | | Deposit date: | 2014-11-24 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Greater Affinity of JC Polyomavirus Capsid for alpha 2,6-Linked Lactoseries Tetrasaccharide c than for Other Sialylated Glycans Is a Major Determinant of Infectivity.
J.Virol., 89, 2015
|
|
8FG8
 
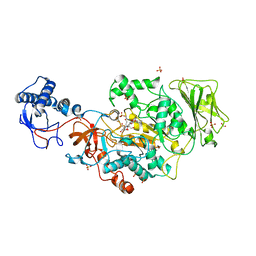 | | Catalytic domain of GtfB in complex with inhibitor 2-[(2,4,5-Trihydroxyphenyl)methylidene]-1-benzofuran-3-one | | Descriptor: | (2Z)-2-[(2,4,5-trihydroxyphenyl)methylidene]-1-benzofuran-3(2H)-one, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Schormann, N, Deivanayagam, C, Velu, S. | | Deposit date: | 2022-12-12 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Hydrogel-Encapsulated Biofilm Inhibitors Abrogate the Cariogenic Activity of Streptococcus mutans .
J.Med.Chem., 66, 2023
|
|
8RJ7
 
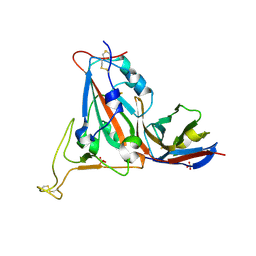 | |
6WRO
 
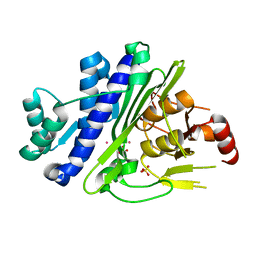 | |
6QQR
 
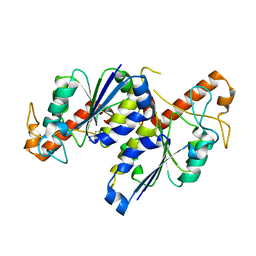 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with inhibitor | | Descriptor: | 1~{H}-indol-5-ylboronic acid, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-19 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Development of Inhibitors againstMycobacterium abscessustRNA (m1G37) Methyltransferase (TrmD) Using Fragment-Based Approaches.
J.Med.Chem., 62, 2019
|
|
8HRT
 
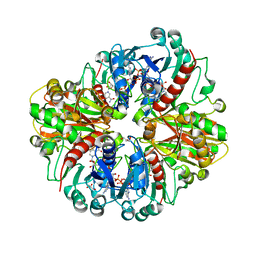 | |
6QMD
 
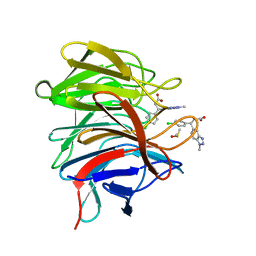 | | Small molecule inhibitor of the KEAP1-NRF2 protein-protein interaction | | Descriptor: | (3~{R})-3-(4-chlorophenyl)-3-(1-methylbenzotriazol-5-yl)propanoic acid, DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1 | | Authors: | Davies, T.G. | | Deposit date: | 2019-02-07 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure-Activity and Structure-Conformation Relationships of Aryl Propionic Acid Inhibitors of the Kelch-like ECH-Associated Protein 1/Nuclear Factor Erythroid 2-Related Factor 2 (KEAP1/NRF2) Protein-Protein Interaction.
J.Med.Chem., 62, 2019
|
|
5X9X
 
 | |
7RT2
 
 | | Crystal Structure of KRAS G12D with compound 25 (4-(4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2R,4R,7aS)-2-fluorotetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}pyrido[4,3-d]pyrimidin-7-yl)naphthalen-2-ol) bound | | Descriptor: | 1,2-ETHANEDIOL, 4-(4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2R,4R,7aS)-2-fluorotetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}pyrido[4,3-d]pyrimidin-7-yl)naphthalen-2-ol, GLYCEROL, ... | | Authors: | Gunn, R.J, Thomas, N.C, Xiaolun, W, Lawson, J.D, Marx, M.A. | | Deposit date: | 2021-08-12 | | Release date: | 2021-12-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Identification of MRTX1133, a Noncovalent, Potent, and Selective KRAS G12D Inhibitor.
J.Med.Chem., 65, 2022
|
|
8ERH
 
 | |
6WLC
 
 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with Uridine-5'-Monophosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Kim, Y, Maltseva, N, Jedrzejczak, R, Endres, M, Chang, C, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-19 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Commun Biol, 4, 2021
|
|
8RI4
 
 | | Crystal structure of the SARS-CoV-2 Main Protease inhibited by (2-methylsulfanyl-6,7-dihydro-[1,4]dioxino[2,3-f]benzimidazol-3-yl)-(p-tolyl)methanone | | Descriptor: | 3C-like proteinase nsp5, 4-METHYLBENZOIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Charton, J, Deprez, B, Hanoulle, X. | | Deposit date: | 2023-12-18 | | Release date: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the SARS-CoV-2 Main Protease inhibited by (2-methylsulfanyl-6,7-dihydro-[1,4]dioxino[2,3-f]benzimidazol-3-yl)-(p-tolyl)methanone
To Be Published
|
|
6QME
 
 | | Small molecule inhibitor of the KEAP1-NRF2 protein-protein interaction | | Descriptor: | (3~{S})-3-(4-chloranyl-3-methyl-phenyl)-3-(1-methylbenzotriazol-5-yl)propanoic acid, Kelch-like ECH-associated protein 1, SULFATE ION | | Authors: | Davies, T.G. | | Deposit date: | 2019-02-07 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure-Activity and Structure-Conformation Relationships of Aryl Propionic Acid Inhibitors of the Kelch-like ECH-Associated Protein 1/Nuclear Factor Erythroid 2-Related Factor 2 (KEAP1/NRF2) Protein-Protein Interaction.
J.Med.Chem., 62, 2019
|
|
1AXU
 
 | | SOLUTION NMR STRUCTURE OF THE [AP]DG ADDUCT OPPOSITE DA IN A DNA DUPLEX, NMR, 9 STRUCTURES | | Descriptor: | DNA DUPLEX D(CCATC-[AP]G-CTACC)D(GGTAGAGATGG), N-1-AMINOPYRENE | | Authors: | Gu, Z, Gorin, A.A, Krishnasami, R, Hingerty, B.E, Basu, A.K, Broyde, S, Patel, D.J. | | Deposit date: | 1997-10-21 | | Release date: | 1998-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-(deoxyguanosin-8-yl)-1-aminopyrene ([AP]dG) adduct opposite dA in a DNA duplex.
Biochemistry, 38, 1999
|
|
5NHK
 
 | | Structure of Ferric uptake regulator from francisella tularensis with Iron | | Descriptor: | FE (III) ION, Ferric uptake regulation protein, ZINC ION | | Authors: | Perard, J, Arnaud, L, Carpentier, P, Michaud-Soret, I, Cavazza, C. | | Deposit date: | 2017-03-21 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional studies of the metalloregulator Fur identify a promoter-binding mechanism and its role inFrancisella tularensisvirulence.
Commun Biol, 1, 2018
|
|
4XC1
 
 | | Crystal structure of human 4E10 Fab in complex with its peptide epitope on HIV-1 GP41: crystals cryoprotected with sn-Glycerol 3-phosphate | | Descriptor: | 4E10 FAB HEAVY CHAIN, 4E10 FAB LIGHT CHAIN, CYCLOHEXYLAMMONIUM ION, ... | | Authors: | Irimia, A, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2014-12-17 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystallographic Identification of Lipid as an Integral Component of the Epitope of HIV Broadly Neutralizing Antibody 4E10.
Immunity, 44, 2016
|
|
6XVP
 
 | | Crystal structure of Neprilysin in complex with Sampatrilat. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Neprilysin, ... | | Authors: | Cozier, G.E, Acharya, K.R, Sharma, U. | | Deposit date: | 2020-01-22 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Molecular Basis for Omapatrilat and Sampatrilat Binding to Neprilysin-Implications for Dual Inhibitor Design with Angiotensin-Converting Enzyme.
J.Med.Chem., 63, 2020
|
|
6OS2
 
 | | Structure of synthetic nanobody-stabilized angiotensin II type 1 receptor bound to TRV026 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Wingler, L.M, Staus, D.P, Skiba, M.A, McMahon, C, Kleinhenz, A.L.W, Lefkowitz, R.J, Kruse, A.C. | | Deposit date: | 2019-05-01 | | Release date: | 2020-02-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Angiotensin and biased analogs induce structurally distinct active conformations within a GPCR.
Science, 367, 2020
|
|
9KNH
 
 | | Structural complex of FTO bound with Dac590 | | Descriptor: | 2-[[2-chloranyl-4-(3,5-dimethyl-1,2-oxazol-4-yl)-6-fluoranyl-phenyl]amino]-5-methoxy-benzoic acid, Alpha-ketoglutarate-dependent dioxygenase FTO, FE (III) ION, ... | | Authors: | Yang, C.G, Yang, T. | | Deposit date: | 2024-11-18 | | Release date: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Development of Orally Bioavailable FTO Inhibitors with Potent Antileukemia Efficacy.
J.Med.Chem., 68, 2025
|
|
4XCP
 
 | | Fatty Acid and Retinol binding protein Na-FAR-1 from Necator americanus | | Descriptor: | Nematode fatty acid retinoid binding protein, PALMITIC ACID | | Authors: | Gabrielsen, M, Rey-Burusco, M.F, Ibanez-Shimabukuro, M, Griffiths, K, Kennedy, M.W, Corsico, B, Smith, B.O. | | Deposit date: | 2014-12-18 | | Release date: | 2015-09-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Diversity in the structures and ligand-binding sites of nematode fatty acid and retinol-binding proteins revealed by Na-FAR-1 from Necator americanus.
Biochem.J., 471, 2015
|
|
6WRR
 
 | |
7MFN
 
 | |
8TUC
 
 | | Unphosphorylated CaMKK2 in complex with CC-8977 | | Descriptor: | (4M)-2-cyclopentyl-4-(7-ethoxyquinazolin-4-yl)benzoic acid, 1,2-ETHANEDIOL, Calcium/calmodulin-dependent protein kinase kinase 2, ... | | Authors: | Bernard, S.M, Shanmugasundaram, V, D'Agostino, L. | | Deposit date: | 2023-08-16 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of Small Molecule Inhibitors and Ligand Directed Degraders of Calcium/Calmodulin Dependent Protein Kinase Kinase 1 and 2 (CaMKK1/2).
J.Med.Chem., 66, 2023
|
|
6QU9
 
 | | Fab fragment of an antibody that inhibits polymerisation of alpha-1-antitrypsin | | Descriptor: | FAB 4B12 heavy chain, FAB 4B12 light chain, GLYCEROL, ... | | Authors: | Jagger, A.M, Heyer-Chauhan, N, Lomas, D.A, Irving, J.A. | | Deposit date: | 2019-02-26 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis for Z alpha 1 -antitrypsin polymerization in the liver.
Sci Adv, 6, 2020
|
|
