4L9O
 
 | | Crystal Structure of the Sec13-Sec16 blade-inserted complex from Pichia pastoris | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | McMahon, C, Jeffrey, P.D, Hughson, F.M. | | Deposit date: | 2013-06-18 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Sec16 influences transitional ER sites by regulating rather than organizing COPII.
Mol Biol Cell, 24, 2013
|
|
5VNW
 
 | |
4H5J
 
 | |
4H5I
 
 | |
5VNV
 
 | | Crystal structure of Nb.b201 | | Descriptor: | FORMIC ACID, Nb.b201, PENTAETHYLENE GLYCOL, ... | | Authors: | Kruse, A.C, McMahon, C. | | Deposit date: | 2017-05-01 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Yeast surface display platform for rapid discovery of conformationally selective nanobodies.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DO1
 
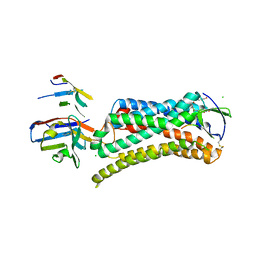 | | Structure of nanobody-stabilized angiotensin II type 1 receptor bound to S1I8 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-like peptide S1I8, ... | | Authors: | Wingler, L.M, McMahon, C, Staus, D.P, Lefkowitz, R.J, Kruse, A.C. | | Deposit date: | 2018-06-08 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Distinctive Activation Mechanism for Angiotensin Receptor Revealed by a Synthetic Nanobody.
Cell, 176, 2019
|
|
6OS0
 
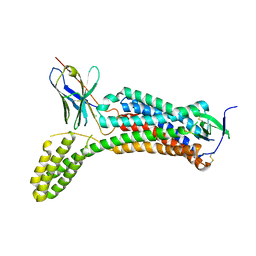 | | Structure of synthetic nanobody-stabilized angiotensin II type 1 receptor bound to angiotensin II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensinogen, CHLORIDE ION, ... | | Authors: | Wingler, L.M, Staus, D.P, Skiba, M.A, McMahon, C, Kleinhenz, A.L.W, Lefkowitz, R.J, Kruse, A.C. | | Deposit date: | 2019-05-01 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Angiotensin and biased analogs induce structurally distinct active conformations within a GPCR.
Science, 367, 2020
|
|
6OS1
 
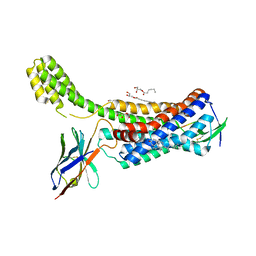 | | Structure of synthetic nanobody-stabilized angiotensin II type 1 receptor bound to TRV023 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHOLESTEROL, Nanobody Nb.AT110i1_le, ... | | Authors: | Wingler, L.M, Staus, D.P, Skiba, M.A, McMahon, C, Kleinhenz, A.L.W, Lefkowitz, R.J, Kruse, A.C. | | Deposit date: | 2019-05-01 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Angiotensin and biased analogs induce structurally distinct active conformations within a GPCR.
Science, 367, 2020
|
|
6OS2
 
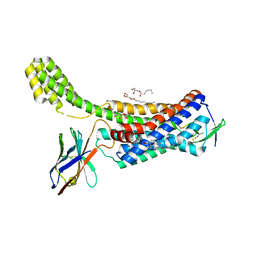 | | Structure of synthetic nanobody-stabilized angiotensin II type 1 receptor bound to TRV026 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Wingler, L.M, Staus, D.P, Skiba, M.A, McMahon, C, Kleinhenz, A.L.W, Lefkowitz, R.J, Kruse, A.C. | | Deposit date: | 2019-05-01 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Angiotensin and biased analogs induce structurally distinct active conformations within a GPCR.
Science, 367, 2020
|
|
