7FIU
 
 | | Crystal structure of the DUB domain of Wolbachia cytoplasmic incompatibility factor CidB from wMel | | Descriptor: | ULP_PROTEASE domain-containing protein | | Authors: | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
7FIT
 
 | | Crystal structure of Wolbachia cytoplasmic incompatibility factor CidA from wMel | | Descriptor: | bacteria factor 1 | | Authors: | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
5AKD
 
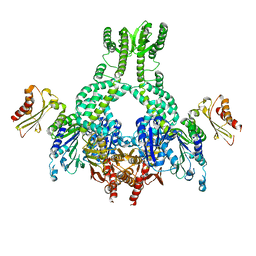 | | MutS in complex with the N-terminal domain of MutL - crystal form 3 | | Descriptor: | DNA MISMATCH REPAIR PROTEIN MUTL, DNA MISMATCH REPAIR PROTEIN MUTS, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Groothuizen, F.S, Winkler, I, Cristovao, M, Fish, A, Winterwerp, H.H.K, Reumer, A, Marx, A.D, Hermans, N, Nicholls, R.A, Murshudov, G.N, Lebbink, J.H.G, Friedhoff, P, Sixma, T.K. | | Deposit date: | 2015-03-03 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (7.6 Å) | | Cite: | MutS/MutL crystal structure reveals that the MutS sliding clamp loads MutL onto DNA.
Elife, 4, 2015
|
|
5AHM
 
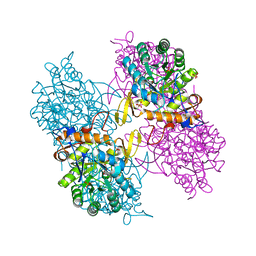 | | IMP-bound form of the DeltaCBS mutant of IMPDH from Pseudomonas aeruginosa | | Descriptor: | INOSINE-5'-MONOPHOSPHATE DEHYDROGENASE, INOSINIC ACID, PHOSPHATE ION | | Authors: | Labesse, G, Alexandre, T, Gelin, M, Haouz, A, Munier-Lehmann, H. | | Deposit date: | 2015-02-06 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystallographic Studies of Two Variants of Pseudomonas Aeruginosa Impdh with Impaired Allosteric Regulation
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5N2X
 
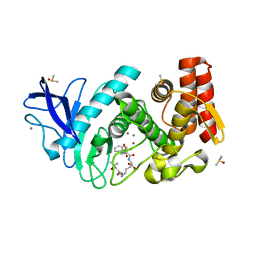 | | Thermolysin in complex with inhibitor JC272 | | Descriptor: | (2~{S})-5-azanyl-2-[[(2~{S})-4-methyl-2-[[oxidanyl(phenylmethoxycarbonylaminomethyl)phosphoryl]amino]pentanoyl]amino]pentanoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Cramer, J, Krimmer, S.G, Heine, A, Klebe, G. | | Deposit date: | 2017-02-08 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.209 Å) | | Cite: | Paying the Price of Desolvation in Solvent-Exposed Protein Pockets: Impact of Distal Solubilizing Groups on Affinity and Binding Thermodynamics in a Series of Thermolysin Inhibitors.
J. Med. Chem., 60, 2017
|
|
7SPD
 
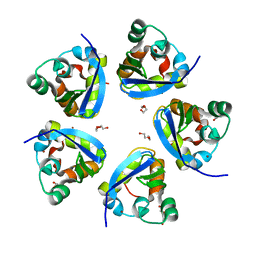 | |
6YXE
 
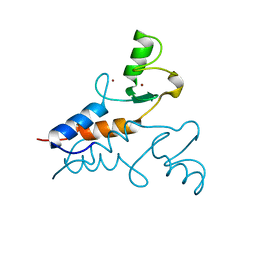 | | Structure of the Trim69 RING domain | | Descriptor: | E3 ubiquitin-protein ligase TRIM69, ZINC ION | | Authors: | Keown, J.R, Goldstone, D.C. | | Deposit date: | 2020-05-01 | | Release date: | 2020-10-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The RING domain of TRIM69 promotes higher-order assembly.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6Z1N
 
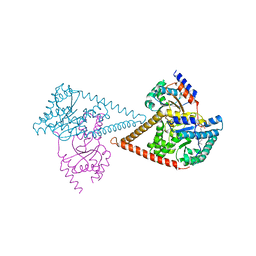 | | Structure of the human heterotetrameric cis-prenyltransferase complex | | Descriptor: | Dehydrodolichyl diphosphate synthase complex subunit DHDDS, Dehydrodolichyl diphosphate synthase complex subunit NUS1, FARNESYL DIPHOSPHATE, ... | | Authors: | Lisnyansky Bar-El, M, Haitin, Y, Giladi, M. | | Deposit date: | 2020-05-14 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of heterotetrameric assembly and disease mutations in the human cis-prenyltransferase complex.
Nat Commun, 11, 2020
|
|
7JPP
 
 | |
5L2R
 
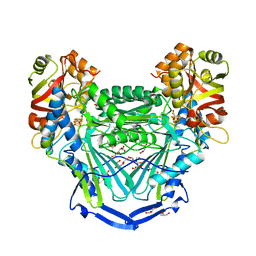 | | Crystal structure of fumarate hydratase from Leishmania major | | Descriptor: | (2S)-2-hydroxybutanedioic acid, DI(HYDROXYETHYL)ETHER, Fumarate hydratase, ... | | Authors: | Feliciano, P.R, Drennan, C.L, Nonato, M.C. | | Deposit date: | 2016-08-02 | | Release date: | 2016-08-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.054 Å) | | Cite: | Crystal structure of an Fe-S cluster-containing fumarate hydratase enzyme from Leishmania major reveals a unique protein fold.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7VMC
 
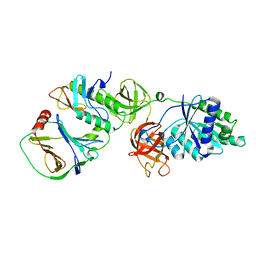 | | Crystal structure of EF-Tu/CdiA/CdiI | | Descriptor: | Contact-dependent inhibitor I, Elongation factor Tu, tRNA nuclease CdiA | | Authors: | Wang, J, Yashiro, Y, Tomita, K. | | Deposit date: | 2021-10-08 | | Release date: | 2022-03-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.413 Å) | | Cite: | Mechanistic insights into tRNA cleavage by a contact-dependent growth inhibitor protein and translation factors.
Nucleic Acids Res., 50, 2022
|
|
7JOA
 
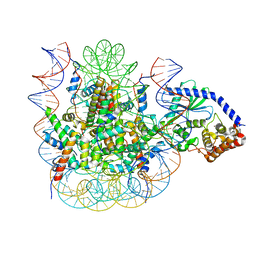 | | 2:1 cGAS-nucleosome complex | | Descriptor: | Cyclic GMP-AMP synthase, DNA (145-MER), Histone H2A type 1, ... | | Authors: | Boyer, J.A, Spangler, C.J, Strauss, J.D, Cesmat, A.P, Liu, P, McGinty, R.K, Zhang, Q. | | Deposit date: | 2020-08-06 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of nucleosome-dependent cGAS inhibition.
Science, 370, 2020
|
|
5VMN
 
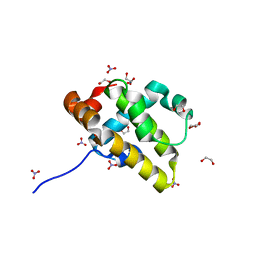 | | Crystal structure of grouper iridovirus GIV66 | | Descriptor: | 1,2-ETHANEDIOL, Bak protein, NITRATE ION | | Authors: | Banjara, S, Kvansakul, M. | | Deposit date: | 2017-04-27 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Grouper iridovirus GIV66 is a Bcl-2 protein that inhibits apoptosis by exclusively sequestering Bim.
J. Biol. Chem., 293, 2018
|
|
5VEX
 
 | | Structure of the human GLP-1 receptor complex with NNC0640 | | Descriptor: | 4-{[(4-cyclohexylphenyl){[3-(methylsulfonyl)phenyl]carbamoyl}amino]methyl}-N-(1H-tetrazol-5-yl)benzamide, Glucagon-like peptide 1 receptor, Endolysin chimera | | Authors: | Song, G, Yang, D, Wang, Y, Graaf, C.D, Zhou, Q, Jiang, S, Liu, K, Cai, X, Dai, A, Lin, G, Liu, D, Wu, F, Wu, Y, Zhao, S, Ye, L, Han, G.W, Lau, J, Wu, B, Hanson, M.A, Liu, Z.-J, Wang, M.-W, Stevens, R.C. | | Deposit date: | 2017-04-05 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human GLP-1 receptor transmembrane domain structure in complex with allosteric modulators.
Nature, 546, 2017
|
|
6WCW
 
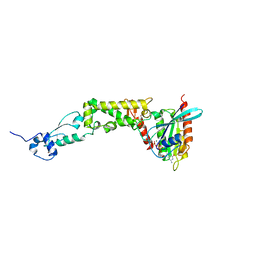 | | Structure of human Rubicon RH domain in complex with GTP-bound Rab7 | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-7a, ... | | Authors: | Bhargava, H.K, Byck, J.M, Farrell, D.P, Anishchenko, I, DiMaio, F, Im, Y.J, Hurley, J.H. | | Deposit date: | 2020-03-31 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for autophagy inhibition by the human Rubicon-Rab7 complex.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8UF7
 
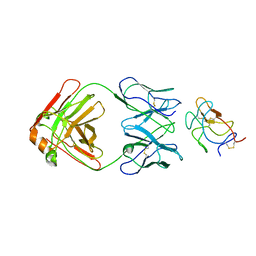 | | Cryo-EM structure of POmAb, a Type-I anti-prothrombin antiphospholipid antibody, bound to kringle-1 of human prothrombin | | Descriptor: | POmAb Heavy Chain, POmAb Light Chain, Prothrombin | | Authors: | Kumar, S, Summers, B, Basore, K, Pozzi, N. | | Deposit date: | 2023-10-03 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure and functional basis of prothrombin recognition by a type I antiprothrombin antiphospholipid antibody.
Blood, 143, 2024
|
|
7EPT
 
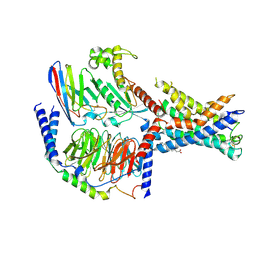 | | Structural basis for the tethered peptide activation of adhesion GPCRs | | Descriptor: | Adhesion G-protein coupled receptor D1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Ping, Y.-Q, Xiao, P, Yang, F, Zhao, R.-J, Guo, S.-C, Yan, X, Wu, X, Liebscher, I, Xu, H.E, Sun, J.-P. | | Deposit date: | 2021-04-27 | | Release date: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for the tethered peptide activation of adhesion GPCRs.
Nature, 604, 2022
|
|
7EQ1
 
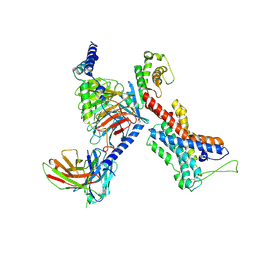 | | GPR114-Gs-scFv16 complex | | Descriptor: | Adhesion G-protein coupled receptor G5, Gs protein alpha subunit, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ping, Y. | | Deposit date: | 2021-04-28 | | Release date: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for the tethered peptide activation of adhesion GPCRs.
Nature, 604, 2022
|
|
8U7Q
 
 | | Human retinal variant phosphomimetic IMPDH1(546)-S477D filament bound by GTP, ATP, IMP, and NAD+, octamer-centered | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | Calise, S.J, Kollman, J.M. | | Deposit date: | 2023-09-15 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Light-sensitive phosphorylation regulates retinal IMPDH1 activity and filament assembly.
J.Cell Biol., 223, 2024
|
|
5L0J
 
 | |
8URN
 
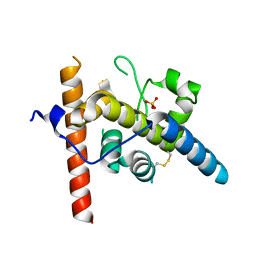 | | Crystal structure of EscI(51-87)-linker-EtgA(18-152) fusion protein | | Descriptor: | EscI inner rod protein type III secretion system,EtgA protein, SULFATE ION | | Authors: | van den Akker, F. | | Deposit date: | 2023-10-26 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural insights into peptidoglycan glycosidase EtgA binding to the inner rod protein EscI of the type III secretion system via a designed EscI-EtgA fusion protein.
Protein Sci., 33, 2024
|
|
7W0N
 
 | | Cryo-EM structure of a dimeric GPCR-Gi complex with peptide | | Descriptor: | Apelin receptor early endogenous ligand, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, F, Yue, Y, Wu, L.J, Liu, L.E, Hanson, M. | | Deposit date: | 2021-11-18 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (4.21 Å) | | Cite: | Structural insight into apelin receptor-G protein stoichiometry.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7K66
 
 | | Structure of Blood Coagulation Factor VIII in Complex with an Anti-C1 Domain Pathogenic Antibody Inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2A9 heavy chain, ... | | Authors: | Childers, K.C, Gish, J, Jarvis, L, Peters, S, Garrels, C, Smith, I.W, Spencer, H.T, Spiegel, P.C. | | Deposit date: | 2020-09-18 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.92 Å) | | Cite: | Structure of blood coagulation factor VIII in complex with an anti-C1 domain pathogenic antibody inhibitor.
Blood, 137, 2021
|
|
7W0M
 
 | | Cryo-EM structure of a monomeric GPCR-Gi complex with small molecule | | Descriptor: | (1R,2S)-N-[4-(2,6-dimethoxyphenyl)-5-(6-methylpyridin-2-yl)-1,2,4-triazol-3-yl]-1-(5-methylpyrimidin-2-yl)-1-oxidanyl-propane-2-sulfonamide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, F, Yue, Y, Liu, L.E, Wu, L.J, Hanson, M. | | Deposit date: | 2021-11-18 | | Release date: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structural insight into apelin receptor-G protein stoichiometry.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5VJ9
 
 | |
