5TFZ
 
 | |
5TG0
 
 | |
5V7Y
 
 | |
5HV0
 
 | |
5HV4
 
 | |
5IAX
 
 | |
5IAT
 
 | |
5IAV
 
 | |
7UI7
 
 | |
7JXV
 
 | | ANTH domain of CALM (clathrin-assembly lymphoid myeloid leukemia protein) bound to ubiquitin | | Descriptor: | Phosphatidylinositol-binding clathrin assembly protein, Ubiquitin | | Authors: | Pashkova, N, Gakhar, L, Schnicker, N.J, Piper, R.C. | | Deposit date: | 2020-08-28 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | ANTH domains within CALM, HIP1R, and Sla2 recognize ubiquitin internalization signals.
Elife, 10, 2021
|
|
7S16
 
 | | Crystal structure of alpha-COP-WD40 domain R57A mutant | | Descriptor: | Coatomer subunit alpha, SODIUM ION | | Authors: | Dey, D, Singh, S, Khan, S, Martin, M, Schnicker, N, Gakhar, L, Pierce, B, Hasan, S.S. | | Deposit date: | 2021-09-01 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | An extended motif in the SARS-CoV-2 spike modulates binding and release of host coatomer in retrograde trafficking
Commun Biol, 5, 2022
|
|
7S22
 
 | | Crystal structure of alpha-COP-WD40 domain | | Descriptor: | Coatomer subunit alpha | | Authors: | Dey, D, Singh, S, Khan, S, Martin, M, Schnicker, N, Gakhar, L, Pierce, B, Hasan, S.S. | | Deposit date: | 2021-09-02 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An extended motif in the SARS-CoV-2 spike modulates binding and release of host coatomer in retrograde trafficking
Commun Biol, 5, 2022
|
|
7S23
 
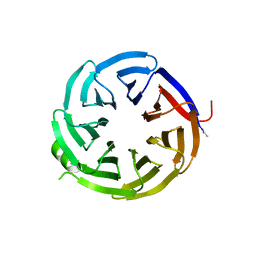 | | Crystal structure of alpha-COP-WD40 domain, Y139A mutant | | Descriptor: | Coatomer subunit alpha | | Authors: | Dey, D, Singh, S, Khan, S, Martin, M, Schnicker, N, Gakhar, L, Pierce, B, Hasan, S.S. | | Deposit date: | 2021-09-03 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | An extended motif in the SARS-CoV-2 spike modulates binding and release of host coatomer in retrograde trafficking
Commun Biol, 5, 2022
|
|
7RE5
 
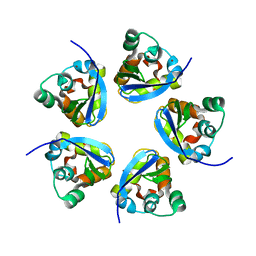 | |
7SPD
 
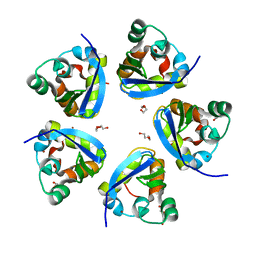 | |
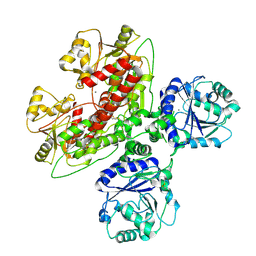
7UI6
 
 | |
