6PG9
 
 | |
2X6D
 
 | | Aurora-A bound to an inhibitor | | Descriptor: | 6-BROMO-7-[4-(4-CHLOROBENZYL)PIPERAZIN-1-YL]-2-[4-(MORPHOLIN-4-YLMETHYL)PHENYL]-3H-IMIDAZO[4,5-B]PYRIDINE, SERINE/THREONINE-PROTEIN KINASE 6, SULFATE ION | | Authors: | Kosmopoulou, M, Bayliss, R. | | Deposit date: | 2010-02-17 | | Release date: | 2010-07-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Imidazo[4,5-b]pyridine derivatives as inhibitors of Aurora kinases: lead optimization studies toward the identification of an orally bioavailable preclinical development candidate.
J. Med. Chem., 53, 2010
|
|
2WP2
 
 | | Structure of Brdt bromodomain BD1 bound to a diacetylated histone H4 peptide. | | Descriptor: | BROMODOMAIN TESTIS-SPECIFIC PROTEIN, HISTONE H4 | | Authors: | Moriniere, J, Rousseaux, S, Steuerwald, U, Soler-Lopez, M, Curtet, S, Vitte, A.-L, Govin, J, Gaucher, J, Sadoul, K, Hart, D.J, Krijgsveld, J, Khochbin, S, Mueller, C.W, Petosa, C. | | Deposit date: | 2009-08-02 | | Release date: | 2009-09-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Cooperative Binding of Two Acetylation Marks on a Histone Tail by a Single Bromodomain.
Nature, 461, 2009
|
|
1E32
 
 | | Structure of the N-Terminal domain and the D1 AAA domain of membrane fusion ATPase p97 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, P97 | | Authors: | Zhang, X, Shaw, A, Bates, P.A, Gorman, M.A, Kondo, H, Dokurno, P, Leonard M, G, Sternberg, J.E, Freemont, P.S. | | Deposit date: | 2000-06-05 | | Release date: | 2001-05-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the Aaa ATPase P97
Mol.Cell, 6, 2000
|
|
6PGA
 
 | |
6PAI
 
 | |
6PPA
 
 | | Crystal structure of the unliganded bromodomain of human BRD7 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 7, PHOSPHATE ION | | Authors: | Chan, A, Karim, M.R, Zhu, J, Schonbrunn, E. | | Deposit date: | 2019-07-05 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
2X2A
 
 | | Free acetyl-CypA trigonal form | | Descriptor: | GLYCEROL, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A, SULFATE ION | | Authors: | Lammers, M, Neumann, H, Chin, J.W, James, L.C. | | Deposit date: | 2010-01-12 | | Release date: | 2010-03-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Acetylation Regulates Cyclophilin a Catalysis, Immunosuppression and HIV Isomerization.
Nat.Chem.Biol., 6, 2010
|
|
6PF5
 
 | | Crystal structure of human thymidylate synthase Delta (7-29) in complex with dUMP and 2-(2-(4-((2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl)benzamido)-4-methoxyphenyl)acetic acid | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Thymidylate synthase, [2-({4-[(2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl]benzene-1-carbonyl}amino)-4-methoxyphenyl]acetic acid | | Authors: | Czyzyk, D.J, Valhondo, M, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2019-06-21 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure activity relationship towards design of cryptosporidium specific thymidylate synthase inhibitors.
Eur.J.Med.Chem., 183, 2019
|
|
6PLF
 
 | | Crystal structure of human PHGDH complexed with Compound 1 | | Descriptor: | 1,2-ETHANEDIOL, 4-{(1S)-1-[(5-chloro-6-{[(5S)-2-oxo-1,3-oxazolidin-5-yl]methoxy}-1H-indole-2-carbonyl)amino]-2-hydroxyethyl}benzoic acid, D-3-phosphoglycerate dehydrogenase | | Authors: | Olland, A, Lakshminarasimhan, D, White, A, Suto, R.K. | | Deposit date: | 2019-06-30 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inhibition of 3-phosphoglycerate dehydrogenase (PHGDH) by indole amides abrogates de novo serine synthesis in cancer cells.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
2XAQ
 
 | | Crystal structure of LSD1-CoREST in complex with a tranylcypromine derivative (MC2584, 13b) | | Descriptor: | 3-{4-[(PHENYLCARBONYL)AMINO]PHENYL}PROPANOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1, ... | | Authors: | Binda, C, Valente, S, Romanenghi, M, Pilotto, S, Cirilli, R, Karytinos, A, Ciossani, G, Botrugno, O.A, Forneris, F, Tardugno, M, Edmondson, D.E, Minucci, S, Mattevi, A, Mai, A. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Biochemical, Structural, and Biological Evaluation of Tranylcypromine Derivatives as Inhibitors of Histone Demethylases Lsd1 and Lsd2.
J.Am.Chem.Soc., 132, 2010
|
|
6Q0W
 
 | | Structure of DDB1-DDA1-DCAF15 complex bound to Indisulam and RBM39 | | Descriptor: | DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, DNA damage-binding protein 1, ... | | Authors: | Faust, T, Yoon, H, Nowak, R.P, Donovan, K.A, Li, Z, Cai, Q, Eleuteri, N.A, Zhang, T, Gray, N.S, Fischer, E.S. | | Deposit date: | 2019-08-02 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural complementarity facilitates E7820-mediated degradation of RBM39 by DCAF15.
Nat.Chem.Biol., 16, 2020
|
|
2ZKE
 
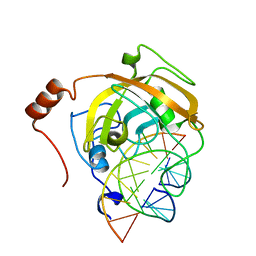 | | Crystal structure of the SRA domain of mouse Np95 in complex with hemi-methylated CpG DNA | | Descriptor: | DNA (5'-D(*DCP*DTP*DAP*DCP*DCP*DGP*DGP*DAP*DTP*DTP*DGP*DC)-3'), DNA (5'-D(*DGP*DCP*DAP*DAP*DTP*DCP*(5CM)P*DGP*DGP*DTP*DAP*DG)-3'), E3 ubiquitin-protein ligase UHRF1 | | Authors: | Arita, K, Ariyoshi, M, Tochio, H, Nakamura, Y, Shirakawa, M. | | Deposit date: | 2008-03-19 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism
Nature, 455, 2008
|
|
2ZVK
 
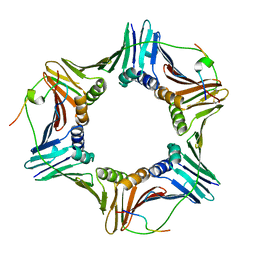 | | Crystal structure of PCNA in complex with DNA polymerase eta fragment | | Descriptor: | DNA polymerase eta, Proliferating cell nuclear antigen | | Authors: | Hishiki, A, Hashimoto, H, Hanafusa, T, Kamei, K, Ohashi, E, Shimizu, T, Ohmori, H, Sato, M. | | Deposit date: | 2008-11-11 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for Novel Interactions between Human Translesion Synthesis Polymerases and Proliferating Cell Nuclear Antigen
J.Biol.Chem., 284, 2009
|
|
6Q00
 
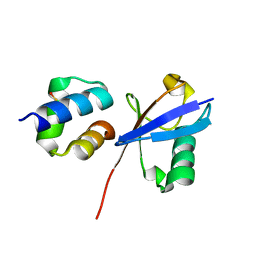 | | TDP2 UBA Domain Bound to Ubiquitin at 0.85 Angstroms Resolution, Crystal Form 1 | | Descriptor: | POTASSIUM ION, Tyrosyl-DNA phosphodiesterase 2, Ubiquitin | | Authors: | Schellenberg, M.J, Krahn, J.M, Williams, R.S. | | Deposit date: | 2019-08-01 | | Release date: | 2020-04-29 | | Last modified: | 2020-06-24 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Ubiquitin stimulated reversal of topoisomerase 2 DNA-protein crosslinks by TDP2.
Nucleic Acids Res., 48, 2020
|
|
1EJF
 
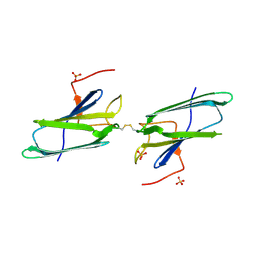 | | CRYSTAL STRUCTURE OF THE HUMAN CO-CHAPERONE P23 | | Descriptor: | Prostaglandin E synthase 3, SULFATE ION | | Authors: | Weaver, A.J, Sullivan, W.P, Felts, S.J, Owen, B.A.L, Toft, D.O. | | Deposit date: | 2000-03-02 | | Release date: | 2000-06-19 | | Last modified: | 2020-10-28 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structure and activity of human p23, a heat shock protein 90 co-chaperone.
J.Biol.Chem., 275, 2000
|
|
6Q3Z
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with the inhibitor 16k | | Descriptor: | (7~{R})-2-[[2-ethoxy-4-(1-methylpiperidin-4-yl)phenyl]amino]-7-ethyl-5-methyl-8-[(4-methylthiophen-2-yl)methyl]-7~{H}-pteridin-6-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Heidenreich, D, Watts, E, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Knapp, S, Hoelder, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-04 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Designing Dual Inhibitors of Anaplastic Lymphoma Kinase (ALK) and Bromodomain-4 (BRD4) by Tuning Kinase Selectivity.
J.Med.Chem., 62, 2019
|
|
2XAG
 
 | | Crystal structure of LSD1-CoREST in complex with para-bromo-(-)-trans- 2-phenylcyclopropyl-1-amine | | Descriptor: | 3-(4-BROMOPHENYL)PROPANAMIDE, FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1, ... | | Authors: | Binda, C, Valente, S, Romanenghi, M, Pilotto, S, Cirilli, R, Karytinos, A, Ciossani, G, Botrugno, O.A, Forneris, F, Tardugno, M, Edmondson, D.E, Minucci, S, Mattevi, A, Mai, A. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Biochemical, Structural, and Biological Evaluation of Tranylcypromine Derivatives as Inhibitors of Histone Demethylases Lsd1 and Lsd2.
J.Am.Chem.Soc., 132, 2010
|
|
2XAJ
 
 | | Crystal structure of LSD1-CoREST in complex with (-)-trans-2- phenylcyclopropyl-1-amine | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1, PHENYLETHYLENECARBOXYLIC ACID, ... | | Authors: | Binda, C, Valente, S, Romanenghi, M, Pilotto, S, Cirilli, R, Karytinos, A, Ciossani, G, Botrugno, O.A, Forneris, F, Tardugno, M, Edmondson, D.E, Minucci, S, Mattevi, A, Mai, A. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Biochemical, Structural, and Biological Evaluation of Tranylcypromine Derivatives as Inhibitors of Histone Demethylases Lsd1 and Lsd2.
J.Am.Chem.Soc., 132, 2010
|
|
6PS9
 
 | | Crystal structure of BRD4 bromodomain 1 with N-methylpyrrolidin-2-one (NMP) derivative 17 (5-{2-[(3R)-1-methyl-5-oxopyrrolidin-3-yl]ethyl}-2,3,4,5-tetrahydro-1H-pyrido[4,3-b]indol-1-one) | | Descriptor: | 5-{2-[(3R)-1-methyl-5-oxopyrrolidin-3-yl]ethyl}-2,3,4,5-tetrahydro-1H-pyrido[4,3-b]indol-1-one, Bromodomain-containing protein 4 | | Authors: | Ilyichova, O.V, Scanlon, M.J. | | Deposit date: | 2019-07-12 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Synthesis and elaboration of N-methylpyrrolidone as an acetamide fragment substitute in bromodomain inhibition.
Bioorg.Med.Chem., 27, 2019
|
|
1BYU
 
 | | CANINE GDP-RAN | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PROTEIN (GTP-BINDING PROTEIN RAN) | | Authors: | Stewart, M, Kent, H.M, Mccoy, A.J. | | Deposit date: | 1998-10-18 | | Release date: | 1998-12-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structure of the Q69L mutant of GDP-Ran shows a major conformational change in the switch II loop that accounts for its failure to bind nuclear transport factor 2 (NTF2).
J.Mol.Biol., 284, 1998
|
|
6PSB
 
 | | Crystal structure of BRD4 bromodomain 1 with N-methylpyrrolidin-2-one (NMP) derivative 18 (5-{[(3R)-1-methyl-5-oxopyrrolidin-3-yl]methyl}-2,3,4,5-tetrahydro-1H-pyrido[4,3-b]indol-1-one) | | Descriptor: | 5-{[(3R)-1-methyl-5-oxopyrrolidin-3-yl]methyl}-2,3,4,5-tetrahydro-1H-pyrido[4,3-b]indol-1-one, Bromodomain-containing protein 4 | | Authors: | Ilyichova, O.V, Scanlon, M.J. | | Deposit date: | 2019-07-12 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Synthesis and elaboration of N-methylpyrrolidone as an acetamide fragment substitute in bromodomain inhibition.
Bioorg.Med.Chem., 27, 2019
|
|
2XL2
 
 | |
2XNE
 
 | | Structure of Aurora-A bound to an imidazopyrazine inhibitor | | Descriptor: | 3-chloro-N-(4-morpholin-4-ylphenyl)-6-pyridin-3-ylimidazo[1,2-a]pyrazin-8-amine, SERINE/THREONINE-PROTEIN KINASE 6 | | Authors: | Kosmopoulou, M, Bayliss, R. | | Deposit date: | 2010-08-02 | | Release date: | 2010-09-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of imidazo[1,2-a]pyrazine derivatives as selective inhibitors of Aurora-A kinase in cells.
Bioorg. Med. Chem. Lett., 20, 2010
|
|
6Q01
 
 | | TDP2 UBA Domain Bound to Ubiquitin at 0.85 Angstroms Resolution, Crystal Form 2 | | Descriptor: | 1,2-ETHANEDIOL, BENZOIC ACID, MAGNESIUM ION, ... | | Authors: | Schellenberg, M.J, Krahn, J.M, Williams, R.S. | | Deposit date: | 2019-08-01 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (0.851 Å) | | Cite: | Ubiquitin stimulated reversal of topoisomerase 2 DNA-protein crosslinks by TDP2.
Nucleic Acids Res., 48, 2020
|
|
