3CNM
 
 | | Crystal Structure of Phenazine Biosynthesis Protein PhzA/B from Burkholderia cepacia R18194, DHHA complex | | Descriptor: | (2S,3S)-TRANS-2,3-DIHYDRO-3-HYDROXYANTHRANILIC ACID, ACETATE ION, Phenazine biosynthesis protein A/B | | Authors: | Ahuja, E.G, Blankenfeldt, W. | | Deposit date: | 2008-03-26 | | Release date: | 2008-12-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | PhzA/B catalyzes the formation of the tricycle in phenazine biosynthesis.
J.Am.Chem.Soc., 130, 2008
|
|
8IBQ
 
 | | Bromodomain and Extra-terminal Domain (BET) BRD4 | | Descriptor: | 7-[2-fluoranyl-3-(1,3,5-trimethylpyrazol-4-yl)phenyl]-1~{H}-imidazo[4,5-b]pyridine, Bromodomain-containing protein 4 | | Authors: | Cao, D, Zhiyan, D, Xiong, B. | | Deposit date: | 2023-02-10 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Discovery of 1 H -Imidazo[4,5- b ]pyridine Derivatives as Potent and Selective BET Inhibitors for the Management of Neuropathic Pain.
J.Med.Chem., 66, 2023
|
|
7S0R
 
 | |
3SDL
 
 | | Crystal structure of human ISG15 in complex with NS1 N-terminal region from influenza B virus, Northeast Structural Genomics Consortium Target IDs HX6481, HR2873, and OR2 | | Descriptor: | Non-structural protein 1, Ubiquitin-like protein ISG15 | | Authors: | Guan, R, Ma, L.-C, Krug, R.M, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-06-09 | | Release date: | 2011-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural basis for the sequence-specific recognition of human ISG15 by the NS1 protein of influenza B virus.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6EW8
 
 | |
6EW6
 
 | | Crystal structure of the BCL6 BTB domain in complex with anilinopyrimidine ligand | | Descriptor: | B-cell lymphoma 6 protein, ~{N}2-(2-chlorophenyl)-1,3,5-triazine-2,4-diamine | | Authors: | Robb, G, Ferguson, A, Hargreaves, D. | | Deposit date: | 2017-11-03 | | Release date: | 2018-10-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Development of a Novel B-Cell Lymphoma 6 (BCL6) PROTAC To Provide Insight into Small Molecule Targeting of BCL6.
ACS Chem. Biol., 13, 2018
|
|
5XEZ
 
 | | Structure of the Full-length glucagon class B G protein-coupled receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-{[(4-cyclohexylphenyl){[3-(methylsulfonyl)phenyl]carbamoyl}amino]methyl}-N-(1H-tetrazol-5-yl)benzamide, ... | | Authors: | Zhang, H, Qiao, A, Yang, D, Yang, L, Dai, A, de Graaf, C, Reedtz-Runge, S, Dharmarajan, V, Zhang, H, Han, G.W, Grant, T, Sierra, R, Weierstall, U, Nelson, G, Liu, W, Wu, Y, Ma, L, Cai, X, Lin, G, Wu, X, Geng, Z, Dong, Y, Song, G, Griffin, P, Lau, J, Cherezov, V, Yang, H, Hanson, M, Stevens, R, Jiang, H, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2017-04-06 | | Release date: | 2017-05-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the full-length glucagon class B G-protein-coupled receptor.
Nature, 546, 2017
|
|
5D6P
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-ethyl-3-[4-(hydroxymethyl)-5-(1H-pyrrol-2-yl)-1,3-thiazol-2-yl]urea, DNA gyrase subunit B, ... | | Authors: | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | Deposit date: | 2015-08-12 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of Azaindole Ureas as a Novel Class of Bacterial Gyrase B Inhibitors.
J.Med.Chem., 58, 2015
|
|
5D6Q
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-ethyl-3-{4-[(E)-2-(pyridin-3-yl)ethenyl]-5-(1H-pyrrol-2-yl)-1,3-thiazol-2-yl}urea, DNA gyrase subunit B, ... | | Authors: | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | Deposit date: | 2015-08-12 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of Azaindole Ureas as a Novel Class of Bacterial Gyrase B Inhibitors.
J.Med.Chem., 58, 2015
|
|
6TPA
 
 | | CDK8/CyclinC in complex with drug ETP-50775 | | Descriptor: | (2~{R})-butane-1,2-diol, 1-[4-chloranyl-3-(trifluoromethyl)phenyl]-3-(5-oxidanylidene-6-pyridin-4-yl-pyrido[2,3-b][1,5]benzoxazepin-9-yl)urea, Cyclin-C, ... | | Authors: | Munoz, I.G, Pastor, J, Martinez, S. | | Deposit date: | 2019-12-12 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Pyrido[2,3-b][1,5]benzoxazepin-5(6H)-one derivatives as CDK8 inhibitors.
Eur.J.Med.Chem., 201, 2020
|
|
4V2O
 
 | | Structure of saposin B in complex with chloroquine | | Descriptor: | N4-(7-CHLORO-QUINOLIN-4-YL)-N1,N1-DIETHYL-PENTANE-1,4-DIAMINE, SAPOSIN-B | | Authors: | Zubieta, C, Lai, X, Doyle, R.P. | | Deposit date: | 2014-10-13 | | Release date: | 2015-12-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | The Lysosomal Protein Saposin B Binds Chloroquine.
Chemmedchem, 11, 2016
|
|
8AU4
 
 | | Structural insights reveal a heterotetramer between oncogenic K-Ras4BG12V and Rgl2, a RalA/B activator | | Descriptor: | Ral guanine nucleotide dissociation stimulator-like 2 | | Authors: | Tariq, M, Ikeya, T, Togashi, N, Fairall, L, Alejo, C.B, Kamei, S, Alonso, B.R, Campillo, M.A.M, Hudson, A, Ito, Y, Schwabe, J, Dominguez, C, Tanaka, K. | | Deposit date: | 2022-08-25 | | Release date: | 2023-08-23 | | Last modified: | 2023-10-25 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the complex of oncogenic KRas4B G12V and Rgl2, a RalA/B activator.
Life Sci Alliance, 7, 2024
|
|
8IDH
 
 | | Bromodomain and Extra-terminal Domain (BET) BRD4 | | Descriptor: | 7-[2-fluoranyl-3-(1,3,5-trimethylpyrazol-4-yl)phenyl]-1~{H}-imidazo[4,5-b]pyridine, Bromodomain-containing protein 4 | | Authors: | Cao, D, Zhiyan, D, Xiong, B. | | Deposit date: | 2023-02-13 | | Release date: | 2023-10-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Discovery of 1 H -Imidazo[4,5- b ]pyridine Derivatives as Potent and Selective BET Inhibitors for the Management of Neuropathic Pain.
J.Med.Chem., 66, 2023
|
|
8BLW
 
 | |
7PZ9
 
 | | HBc-F97L premature secretion phenotype | | Descriptor: | Capsid protein | | Authors: | Makbul, C, Boettcher, B. | | Deposit date: | 2021-10-11 | | Release date: | 2021-12-08 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Binding of a Pocket Factor to Hepatitis B Virus Capsids Changes the Rotamer Conformation of Phenylalanine 97.
Viruses, 13, 2021
|
|
7PZN
 
 | |
7PZI
 
 | |
7PZK
 
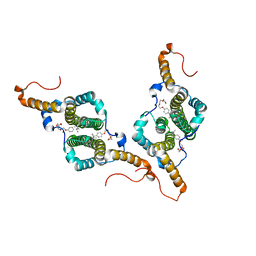 | | HBc-WT in complex with Triton X-100 | | Descriptor: | Capsid protein, FRAGMENT OF TRITON X-100 | | Authors: | Makbul, C, Boettcher, B. | | Deposit date: | 2021-10-12 | | Release date: | 2021-12-08 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Binding of a Pocket Factor to Hepatitis B Virus Capsids Changes the Rotamer Conformation of Phenylalanine 97.
Viruses, 13, 2021
|
|
7PZL
 
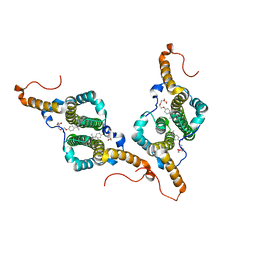 | | HBc-F97L premature secretion phenotype | | Descriptor: | Capsid protein, FRAGMENT OF TRITON X-100 | | Authors: | Makbul, C, Boettcher, B. | | Deposit date: | 2021-10-12 | | Release date: | 2021-12-08 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Binding of a Pocket Factor to Hepatitis B Virus Capsids Changes the Rotamer Conformation of Phenylalanine 97.
Viruses, 13, 2021
|
|
7PZM
 
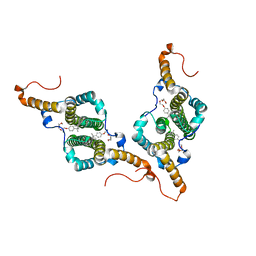 | | HBc-P5T in complex with X-100 | | Descriptor: | Capsid protein, FRAGMENT OF TRITON X-100 | | Authors: | Makbul, C, Boettcher, B. | | Deposit date: | 2021-10-12 | | Release date: | 2021-12-08 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Binding of a Pocket Factor to Hepatitis B Virus Capsids Changes the Rotamer Conformation of Phenylalanine 97.
Viruses, 13, 2021
|
|
7A22
 
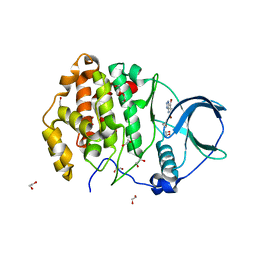 | | Crystal structure of human protein kinase CK2alpha' (CSNK2A2 gene product) in complex with the ATP-competitive inhibitor 5,6,7-tribromo-1H-triazolo[4,5-b]pyridine | | Descriptor: | 1,2-ETHANEDIOL, 5,6,7-tris(bromanyl)-1~{H}-[1,2,3]triazolo[4,5-b]pyridine, Casein kinase II subunit alpha' | | Authors: | Niefind, K, Lindenblatt, D, Toelzer, C, Bretner, M, Chojnacki, K, Wielechowska, M, Winska, P. | | Deposit date: | 2020-08-15 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Synthesis, biological properties and structural study of new halogenated azolo[4,5-b]pyridines as inhibitors of CK2 kinase.
Bioorg.Chem., 106, 2021
|
|
7A2H
 
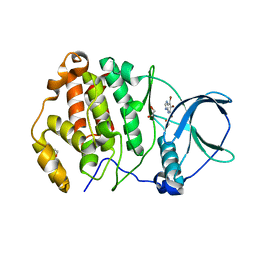 | | Crystal structure of human protein kinase CK2alpha' (CSNK2A2 gene product) in complex with the ATP-competitive inhibitor 5,6,7-tribromo-1H-imidazo[4,5-b]pyridine | | Descriptor: | 1,2-ETHANEDIOL, 5,6,7-tris(bromanyl)-1~{H}-imidazo[4,5-b]pyridine, Casein kinase II subunit alpha' | | Authors: | Niefind, K, Lindenblatt, D, Toelzer, C, Bretner, M, Chojnacki, K, Wielechowska, M, Winska, P. | | Deposit date: | 2020-08-18 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Synthesis, biological properties and structural study of new halogenated azolo[4,5-b]pyridines as inhibitors of CK2 kinase.
Bioorg.Chem., 106, 2021
|
|
6GIL
 
 | | NMR structure of temporin B in SDS micelles | | Descriptor: | Temporin-B | | Authors: | Manzo, G, Mason, J.A. | | Deposit date: | 2018-05-12 | | Release date: | 2018-06-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Minor sequence modifications in temporin B cause drastic changes in antibacterial potency and selectivity by fundamentally altering membrane activity.
Sci Rep, 9, 2019
|
|
6WFS
 
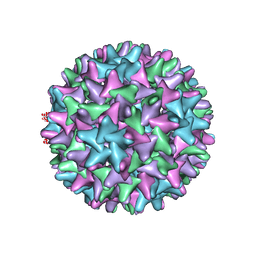 | | Cryo-EM Structure of Hepatitis B virus T=4 capsid in complex with the antiviral molecule DBT1 | | Descriptor: | 11-oxo-N-[2-(4-sulfamoylphenyl)ethyl]-10,11-dihydrodibenzo[b,f][1,4]thiazepine-8-carboxamide, Capsid protein | | Authors: | Schlicksup, C, Laughlin, P, Dunkelbarger, S, Wang, J.C, Zlotnick, A. | | Deposit date: | 2020-04-03 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Local Stabilization of Subunit-Subunit Contacts Causes Global Destabilization of Hepatitis B Virus Capsids.
Acs Chem.Biol., 15, 2020
|
|
8PX6
 
 | |
