1EQY
 
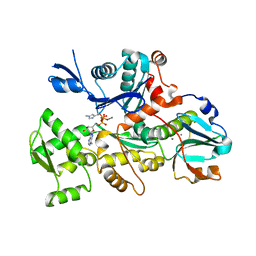 | | COMPLEX BETWEEN RABBIT MUSCLE ALPHA-ACTIN: HUMAN GELSOLIN DOMAIN 1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ALPHA ACTIN, CALCIUM ION, ... | | Authors: | McLaughlin, P.J, Gooch, J.T, Mannherz, H.G, Weeds, A.G. | | Deposit date: | 2000-04-06 | | Release date: | 2000-05-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of gelsolin segment 1-actin complex and the mechanism of filament severing.
Nature, 364, 1993
|
|
6WFS
 
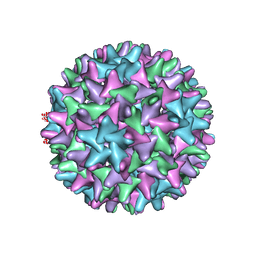 | | Cryo-EM Structure of Hepatitis B virus T=4 capsid in complex with the antiviral molecule DBT1 | | Descriptor: | 11-oxo-N-[2-(4-sulfamoylphenyl)ethyl]-10,11-dihydrodibenzo[b,f][1,4]thiazepine-8-carboxamide, Capsid protein | | Authors: | Schlicksup, C, Laughlin, P, Dunkelbarger, S, Wang, J.C, Zlotnick, A. | | Deposit date: | 2020-04-03 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Local Stabilization of Subunit-Subunit Contacts Causes Global Destabilization of Hepatitis B Virus Capsids.
Acs Chem.Biol., 15, 2020
|
|
1E0B
 
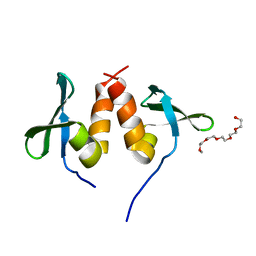 | | Chromo shadow domain from fission yeast swi6 protein. | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, SWI6 PROTEIN | | Authors: | Cowieson, N.P, Partridge, J.F, Allshire, R.C, Mclaughlin, P.J. | | Deposit date: | 2000-03-16 | | Release date: | 2000-05-12 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dimerisation of Chromo Shadow Domain and Distinctions from the Chromodomain as Revealed by Structural Analysis
Curr.Biol., 10, 2000
|
|
2K77
 
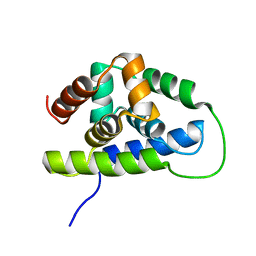 | | NMR solution structure of the Bacillus subtilis ClpC N-domain | | Descriptor: | Negative regulator of genetic competence clpC/mecB | | Authors: | Kojetin, D.J, McLaughlin, P.D, Thompson, R.J, Rance, M, Cavanagh, J. | | Deposit date: | 2008-08-04 | | Release date: | 2009-04-28 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structural and motional contributions of the Bacillus subtilis ClpC N-domain to adaptor protein interactions.
J.Mol.Biol., 387, 2009
|
|
1NPH
 
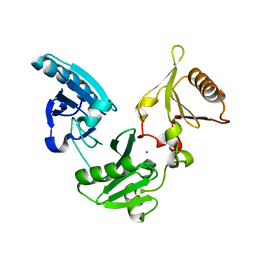 | | Gelsolin Domains 4-6 in Active, Actin Free Conformation Identifies Sites of Regulatory Calcium Ions | | Descriptor: | CALCIUM ION, Gelsolin | | Authors: | Kolappan, S, Gooch, J.T, Weeds, A.G, McLaughlin, P.J. | | Deposit date: | 2003-01-17 | | Release date: | 2003-05-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Gelsolin Domains 4-6 in Active, Actin-Free Conformation Identifies Sites of Regulatory Calcium Ions
J.Mol.Biol., 329, 2003
|
|
1ESV
 
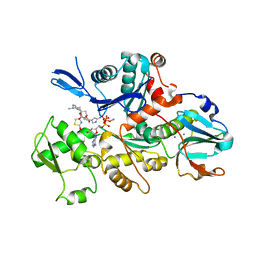 | | COMPLEX BETWEEN LATRUNCULIN A:RABBIT MUSCLE ALPHA ACTIN:HUMAN GELSOLIN DOMAIN 1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ALPHA ACTIN, CALCIUM ION, ... | | Authors: | Morton, W.M, Ayscough, K.A, McLaughlin, P.J. | | Deposit date: | 2000-04-11 | | Release date: | 2000-07-19 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Latrunculin alters the actin-monomer subunit interface to prevent polymerization.
Nat.Cell Biol., 2, 2000
|
|
2JVK
 
 | |
2JVJ
 
 | |
2JVI
 
 | |
