6GZ6
 
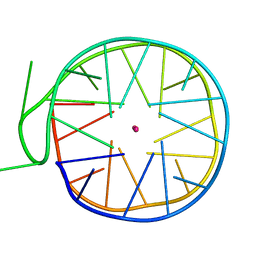 | | Structure of a left-handed G-quadruplex | | Descriptor: | DNA (27-MER), POTASSIUM ION | | Authors: | Bakalar, B, Heddi, B, Schmitt, E, Mechulam, Y, Phan, A.T. | | Deposit date: | 2018-07-03 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | A Minimal Sequence for Left-Handed G-Quadruplex Formation.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
6D37
 
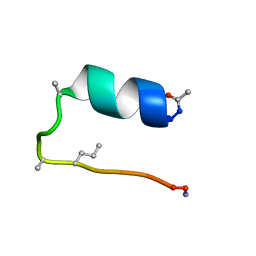 | |
6CVO
 
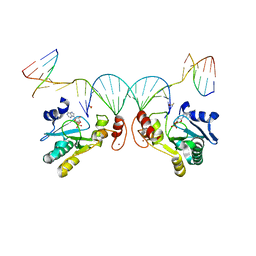 | | Human Aprataxin (Aptx) bound to nicked RNA-DNA, AMP and Zn product complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, Aprataxin, BETA-MERCAPTOETHANOL, ... | | Authors: | Schellenberg, M.J, Tumbale, P.P, Williams, R.S. | | Deposit date: | 2018-03-28 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanism of APTX nicked DNA sensing and pleiotropic inactivation in neurodegenerative disease.
EMBO J., 37, 2018
|
|
6GB1
 
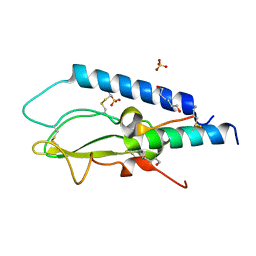 | | Crystal structure of the GLP1 receptor ECD with Peptide 11 | | Descriptor: | Glucagon-like peptide 1 receptor, HEXANE-1,6-DIOL, Peptide 11, ... | | Authors: | Schreuder, H.A, Liesum, A. | | Deposit date: | 2018-04-13 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Dual Glucagon-like Peptide 1 (GLP-1)/Glucagon Receptor Agonists Specifically Optimized for Multidose Formulations.
J. Med. Chem., 61, 2018
|
|
6GBM
 
 | | Solution structure of FUS-RRM bound to stem-loop RNA | | Descriptor: | RNA (5'-R(*GP*GP*CP*AP*GP*AP*UP*UP*AP*CP*AP*AP*UP*UP*CP*UP*AP*UP*UP*UP*GP*CP*C)-3'), RNA-binding protein FUS | | Authors: | Loughlin, F.E, Allain, F.H.-T. | | Deposit date: | 2018-04-15 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of FUS Bound to RNA Reveals a Bipartite Mode of RNA Recognition with Both Sequence and Shape Specificity.
Mol. Cell, 73, 2019
|
|
6CGX
 
 | | Backbone cyclised conotoxin Vc1.1 mutant - D11A, E14A | | Descriptor: | Alpha-conotoxin Vc1A | | Authors: | Clark, R.J. | | Deposit date: | 2018-02-21 | | Release date: | 2018-05-23 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure-Activity Studies Reveal the Molecular Basis for GABAB-Receptor Mediated Inhibition of High Voltage-Activated Calcium Channels by alpha-Conotoxin Vc1.1.
ACS Chem. Biol., 13, 2018
|
|
6COQ
 
 | | CSP1-K6A | | Descriptor: | Competence-stimulating peptide type 1 | | Authors: | Yang, Y. | | Deposit date: | 2018-03-12 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of Competence-Stimulating Peptide Analogues Reveals Key Features for ComD1 and ComD2 Receptor Binding in Streptococcus pneumoniae.
Biochemistry, 57, 2018
|
|
6G4J
 
 | |
6COP
 
 | | CSP1-R3A | | Descriptor: | Competence-stimulating peptide type 1 | | Authors: | Yang, Y. | | Deposit date: | 2018-03-12 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of Competence-Stimulating Peptide Analogues Reveals Key Features for ComD1 and ComD2 Receptor Binding in Streptococcus pneumoniae.
Biochemistry, 57, 2018
|
|
6BZL
 
 | | Solution structure of VEK75 | | Descriptor: | M protein | | Authors: | Qiu, C, Yuan, Y, Castellino, F.J. | | Deposit date: | 2017-12-24 | | Release date: | 2019-01-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Contributions of different modules of the plasminogen-binding Streptococcus pyogenes M-protein that mediate its functional dimerization.
J.Struct.Biol., 204, 2018
|
|
6BPU
 
 | | Crystal structure of ferrous form of the F2-Tyr157 human cysteine dioxygenase with both uncrosslinked and crosslinked cofactor | | Descriptor: | Cysteine dioxygenase type 1, FE (II) ION, GLYCEROL, ... | | Authors: | Liu, A, Li, J, Shin, I. | | Deposit date: | 2017-11-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cleavage of a carbon-fluorine bond by an engineered cysteine dioxygenase.
Nat. Chem. Biol., 14, 2018
|
|
6GWS
 
 | | Crystal structure of human PCNA in complex with three p15 peptides | | Descriptor: | PCNA-associated factor, Proliferating cell nuclear antigen | | Authors: | De March, M, Merino, N, Gonzalez-Magana, A, Romano-Moreno, M, Onesti, S, Blanco, F.J, De Biasio, A. | | Deposit date: | 2018-06-25 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | p15PAF binding to PCNA modulates the DNA sliding surface.
Nucleic Acids Res., 46, 2018
|
|
6D74
 
 | |
6H5T
 
 | | Intersectin SH3A short isoform | | Descriptor: | 2,5,8,11,14,17,20,23-OCTAOXAPENTACOSAN-25-OL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Driller, J.H, Gerth, F, Freund, C, Wahl, M.C, Loll, B. | | Deposit date: | 2018-07-25 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.689 Å) | | Cite: | Exon Inclusion Modulates Conformational Plasticity and Autoinhibition of the Intersectin 1 SH3A Domain.
Structure, 27, 2019
|
|
6GXG
 
 | | Tryparedoxin from Trypanosoma brucei in complex with CFT | | Descriptor: | 5-(4-fluorophenyl)-2-methyl-3~{H}-thieno[2,3-d]pyrimidin-4-one, GLYCEROL, Tryparedoxin | | Authors: | Bader, N, Wagner, A, Hellmich, U, Schindelin, H. | | Deposit date: | 2018-06-27 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Inhibitor-Induced Dimerization of an Essential Oxidoreductase from African Trypanosomes.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
6DFA
 
 | | Kaiso (ZBTB33) E535A zinc finger DNA binding domain in complex with the specific Kaiso binding sequence (KBS) | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*GP*TP*TP*AP*TP*TP*GP*GP*CP*AP*GP*GP*AP*AP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*TP*TP*CP*CP*TP*GP*CP*CP*AP*AP*TP*AP*AP*CP*G)-3'), ... | | Authors: | Nikolova, E.N, Stanfield, R.L, Dyson, H.J, Wright, P.E. | | Deposit date: | 2018-05-14 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.908 Å) | | Cite: | A conformational switch in the zinc finger protein Kaiso mediates differential readout of specific and methylated DNA sequences.
Biochemistry, 2020
|
|
6GXZ
 
 | | Crystal structure of the human RPAP3(TPR2)-PIH1D1(CS) complex | | Descriptor: | CALCIUM ION, PIH1 domain-containing protein 1, RNA polymerase II-associated protein 3 | | Authors: | Henri, J, Quinternet, M, Manival, X, Chagot, M.-E, Charpentier, B, Meyer, P. | | Deposit date: | 2018-06-27 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.965 Å) | | Cite: | Deep Structural Analysis of RPAP3 and PIH1D1, Two Components of the HSP90 Co-chaperone R2TP Complex.
Structure, 26, 2018
|
|
6GIS
 
 | | Structural basis of human clamp sliding on DNA | | Descriptor: | DNA (5'-D(P*AP*TP*AP*CP*GP*AP*TP*GP*GP*G)-3'), DNA (5'-D(P*CP*CP*CP*AP*TP*CP*GP*TP*AP*T)-3'), Proliferating cell nuclear antigen | | Authors: | De March, M, Merino, N, Barrera-Vilarmau, S, Crehuet, R, Onesti, S, Blanco, F.J, De Biasio, A. | | Deposit date: | 2018-05-15 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structural basis of human PCNA sliding on DNA.
Nat Commun, 8, 2017
|
|
6CMS
 
 | | Closed structure of active SHP2 mutant E76K bound to SHP099 inhibitor | | Descriptor: | 6-(4-azanyl-4-methyl-piperidin-1-yl)-3-[2,3-bis(chloranyl)phenyl]pyrazin-2-amine, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Padua, R.A.P, Sun, Y, Marko, I, Pitsawong, W, Kern, D. | | Deposit date: | 2018-03-06 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Mechanism of activating mutations and allosteric drug inhibition of the phosphatase SHP2.
Nat Commun, 9, 2018
|
|
6COT
 
 | | CSP2-d10 | | Descriptor: | Competence-stimulating peptide type 2 | | Authors: | Yang, Y. | | Deposit date: | 2018-03-12 | | Release date: | 2018-08-29 | | Last modified: | 2018-09-19 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of Competence-Stimulating Peptide Analogues Reveals Key Features for ComD1 and ComD2 Receptor Binding in Streptococcus pneumoniae.
Biochemistry, 57, 2018
|
|
6CBI
 
 | | PCNA in complex with inhibitor | | Descriptor: | GLY-ARG-LYS-ARG-ARG-GLN-DAB-SER-MET-THR-GLU-PHE-TYR-HIS, Proliferating cell nuclear antigen, SULFATE ION | | Authors: | Bruning, J.B, Wegener, K.L. | | Deposit date: | 2018-02-03 | | Release date: | 2018-07-04 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Rational Design of a 310-Helical PIP-Box Mimetic Targeting PCNA, the Human Sliding Clamp.
Chemistry, 24, 2018
|
|
6CCI
 
 | | The Crystal Structure of XOAT1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2018-02-07 | | Release date: | 2019-02-20 | | Last modified: | 2020-09-02 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular Mechanism of Polysaccharide Acetylation by the Arabidopsis XylanO-acetyltransferase XOAT1.
Plant Cell, 32, 2020
|
|
6GKG
 
 | |
6AXJ
 
 | |
6AVI
 
 | |
