4WXQ
 
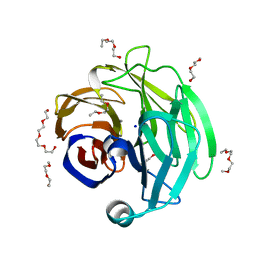 | | Crystal Structure of the Myocilin Olfactomedin Domain | | Descriptor: | CALCIUM ION, HEXAETHYLENE GLYCOL, Myocilin, ... | | Authors: | Orwig, S.D, Turnage, K.C, Donegan, R.K, Lieberman, R.L. | | Deposit date: | 2014-11-14 | | Release date: | 2015-04-01 | | Last modified: | 2017-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for misfolding in myocilin-associated glaucoma.
Hum.Mol.Genet., 24, 2015
|
|
1HZW
 
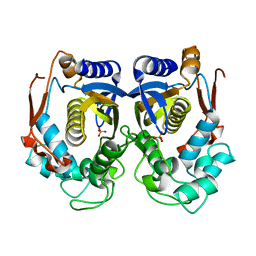 | | CRYSTAL STRUCTURE OF HUMAN THYMIDYLATE SYNTHASE | | Descriptor: | PHOSPHATE ION, THYMIDYLATE SYNTHASE | | Authors: | Almog, R, Waddling, C.A, Maley, F, Maley, G.F, Van Roey, P. | | Deposit date: | 2001-01-26 | | Release date: | 2001-05-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a deletion mutant of human thymidylate synthase Delta (7-29) and its ternary complex with Tomudex and dUMP.
Protein Sci., 10, 2001
|
|
4WY7
 
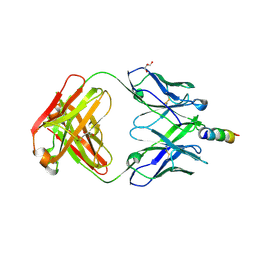 | | Crystal structure of recombinant 4E10 expressed in Escherichia coli with epitope bound | | Descriptor: | Envelope glycoprotein gp160, Fab 4E10 Heavy chain, Fab 4E10Light chain, ... | | Authors: | Rujas, E, Morante, K, Tsumoto, K, Nieva, J.L, Caaveiro, J.M.M. | | Deposit date: | 2014-11-16 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Atomic Structure of the HIV-1 gp41 Transmembrane Domain and Its Connection to the Immunogenic Membrane-proximal External Region.
J.Biol.Chem., 290, 2015
|
|
6I5U
 
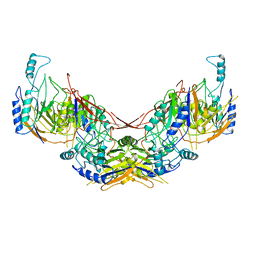 | |
6I80
 
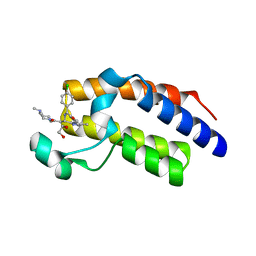 | | Crystal Structure of the second bromodomain of BRD2 in complex with RT53 | | Descriptor: | 1,2-ETHANEDIOL, 2-[(4~{S})-6-(4-chlorophenyl)-8-methoxy-1-methyl-4~{H}-[1,2,4]triazolo[4,3-a][1,4]benzodiazepin-4-yl]-1-[4-(dimethylamino)piperidin-1-yl]ethanone, Bromodomain-containing protein 2 | | Authors: | Picaud, S, Traquete, R, Bernardes, G.J.L, Newman, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Filippakopoulos, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-11-19 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Crystal Structure of the second bromodomain of BRD2 in complex with RT53
To Be Published
|
|
6ECD
 
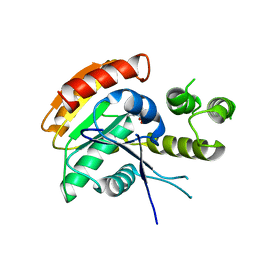 | | Vlm2 thioesterase domain with genetically encoded 2,3-diaminopropionic acid bound with a tetradepsipeptide | | Descriptor: | Vlm2, tetradepsipeptide | | Authors: | Alonzo, D.A, Huguenin-Dezot, N, Heberlig, G.W, Mahesh, M, Nguyen, D.P, Dornan, M.H, Boddy, C.N, Chin, J.W, Schmeing, T.M. | | Deposit date: | 2018-08-07 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Trapping biosynthetic acyl-enzyme intermediates with encoded 2,3-diaminopropionic acid.
Nature, 565, 2019
|
|
6EBV
 
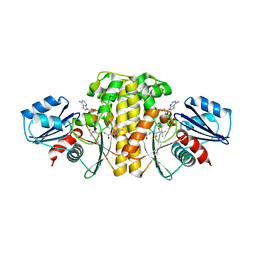 | | Staphylococcus aureus Type II pantothenate kinase in complex with ADP and pantothenate analog Deoxy-N7-Pan | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, N-heptyl-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide, Type II pantothenate kinase | | Authors: | Chen, Y, Antoshchenko, T, Strauss, E, Barnard, L, Huang, Y.H, Park, H. | | Deposit date: | 2018-08-07 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-based identification of uncompetitive inhibitors for Staphylococcus aureus pantothenate kinase.
To Be Published
|
|
4XBC
 
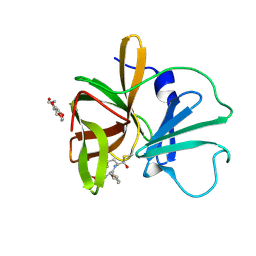 | | 1.60 A resolution structure of Norovirus 3CL protease complex with a covalently bound dipeptidyl inhibitor (1R,2S)-2-({N-[(benzyloxy)carbonyl]-3-cyclohexyl-L-alanyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid (Hexagonal Form) | | Descriptor: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-3-cyclohexyl-L-alanyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-LIKE PROTEASE, TETRAETHYLENE GLYCOL | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Kim, Y, Weerawarna, P.M, Uy, R.A.Z, Damalanka, V.C, Mandadapu, S.R, Alliston, K.R, Groutas, W.C, Chang, K.-O. | | Deposit date: | 2014-12-16 | | Release date: | 2015-03-25 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Design and Optimization of Dipeptidyl Inhibitors of Norovirus 3CL Protease. Structure-Activity Relationships and Biochemical, X-ray Crystallographic, Cell-Based, and In Vivo Studies.
J.Med.Chem., 58, 2015
|
|
4XBB
 
 | | 1.85A resolution structure of Norovirus 3CL protease complex with a covalently bound dipeptidyl inhibitor diethyl [(1R,2S)-2-[(N-{[(3-chlorobenzyl)oxy]carbonyl}-3-cyclohexyl-L-alanyl)amino]-1-hydroxy-3-(2-oxo-2H-pyrrol-3-yl)propyl]phosphonate | | Descriptor: | 3C-LIKE PROTEASE, SULFATE ION, diethyl [(1R,2S)-2-[(N-{[(3-chlorobenzyl)oxy]carbonyl}-3-cyclohexyl-L-alanyl)amino]-1-hydroxy-3-(2-oxo-2H-pyrrol-3-yl)propyl]phosphonate | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Kim, Y, Weerawarna, P.M, Uy, R.A.Z, Damalanka, V.C, Mandadapu, S.R, Alliston, K.R, Groutas, W.C, Chang, K.-O. | | Deposit date: | 2014-12-16 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Guided Design and Optimization of Dipeptidyl Inhibitors of Norovirus 3CL Protease. Structure-Activity Relationships and Biochemical, X-ray Crystallographic, Cell-Based, and In Vivo Studies.
J.Med.Chem., 58, 2015
|
|
6I57
 
 | |
6ELW
 
 | | High resolution structure of selenocysteine containing human GPX4 | | Descriptor: | CHLORIDE ION, Phospholipid hydroperoxide glutathione peroxidase, mitochondrial | | Authors: | Kalms, J, Borchert, A, Kuhn, H, Scheerer, P. | | Deposit date: | 2017-09-29 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure and functional characterization of selenocysteine-containing glutathione peroxidase 4 suggests an alternative mechanism of peroxide reduction.
Biochim. Biophys. Acta, 1863, 2018
|
|
6ECE
 
 | | Vlm2 thioesterase domain with genetically encoded 2,3-diaminopropionic acid bound with a dodecadepsipeptide, space group H3 | | Descriptor: | Vlm2, dodecadepsipeptide | | Authors: | Alonzo, D.A, Huguenin-Dezot, N, Heberlig, G.W, Mahesh, M, Nguyen, D.P, Dornan, M.H, Boddy, C.N, Chin, J.W, Schmeing, T.M. | | Deposit date: | 2018-08-07 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Trapping biosynthetic acyl-enzyme intermediates with encoded 2,3-diaminopropionic acid.
Nature, 565, 2019
|
|
5J5H
 
 | | X-RAY STRUCTURE OF ACETYLCHOLINE BINDING PROTEIN (ACHBP) IN COMPLEX WITH 6-(2-methoxyphenyl)-N4,N4-bis[(pyridin-2-yl)methyl]pyrimidine-2,4-diamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-(2-methoxyphenyl)-N~4~,N~4~-bis[(pyridin-2-yl)methyl]pyrimidine-2,4-diamine, Acetylcholine-binding protein, ... | | Authors: | Kaczanowska, K, Harel, M, Camacho Hernandez, A.G, Taylor, P. | | Deposit date: | 2016-04-02 | | Release date: | 2017-03-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Substituted 2-Aminopyrimidines Selective for alpha 7-Nicotinic Acetylcholine Receptor Activation and Association with Acetylcholine Binding Proteins.
J. Am. Chem. Soc., 139, 2017
|
|
5J5G
 
 | | X-Ray Crystal Structure of Acetylcholine Binding Protein (AChBP) in Complex with 6-(4-methoxyphenyl)-N4,N4-bis[(pyridin-2-yl)methyl]pyrimidine-2,4-diamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-(4-methoxyphenyl)-N~4~,N~4~-bis[(pyridin-2-yl)methyl]pyrimidine-2,4-diamine, Acetylcholine-binding protein, ... | | Authors: | Kaczanowska, K, Harel, M, Camacho Hernandez, A.G, Taylor, P. | | Deposit date: | 2016-04-02 | | Release date: | 2017-03-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.036 Å) | | Cite: | Substituted 2-Aminopyrimidines Selective for alpha 7-Nicotinic Acetylcholine Receptor Activation and Association with Acetylcholine Binding Proteins.
J. Am. Chem. Soc., 139, 2017
|
|
6YNQ
 
 | | Structure of SARS-CoV-2 Main Protease bound to 2-Methyl-1-tetralone. | | Descriptor: | (2~{S})-2-methyl-3,4-dihydro-2~{H}-naphthalen-1-one, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
8CTA
 
 | | Minimal 2:2 Ternary Complex between BI-224436 bound HIV-1 Integrase Catalytic Core Domain Dimer and Carboxy Terminal Domains | | Descriptor: | (2S)-tert-butoxy[4-(2,3-dihydropyrano[4,3,2-de]quinolin-7-yl)-2-methylquinolin-3-yl]acetic acid, 1,2-ETHANEDIOL, Integrase | | Authors: | Gupta, K, Van Duyne, G.D, Eilers, G, Bushman, F.D. | | Deposit date: | 2022-05-13 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Structure of a HIV-1 IN-Allosteric inhibitor complex at 2.93 angstrom resolution: Routes to inhibitor optimization.
Plos Pathog., 19, 2023
|
|
6FK3
 
 | | Structure and function of aldehyde dehydrogenase from Thermus thermophilus: An enzyme with an evolutionarily-distinct C-terminal arm (Recombinant full-length protein in complex with propanal) | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, Aldehyde dehydrogenase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hayes, K.A, Noor, M.R, Djeghader, A, Soulimane, T. | | Deposit date: | 2018-01-23 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The quaternary structure of Thermus thermophilus aldehyde dehydrogenase is stabilized by an evolutionary distinct C-terminal arm extension.
Sci Rep, 8, 2018
|
|
2J3I
 
 | | Crystal structure of Arabidopsis thaliana Double Bond Reductase (AT5G16970)-Binary Complex | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-DEPENDENT OXIDOREDUCTASE P1 | | Authors: | Youn, B, Kim, S.J, Moinuddin, S.G, Lee, C, Bedgar, D.L, Harper, A.R, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2006-08-21 | | Release date: | 2006-10-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanistic and Structural Studies of Apoform, Binary, and Ternary Complexes of the Arabidopsis Alkenal Double Bond Reductase at5G16970.
J.Biol.Chem., 281, 2006
|
|
6EYU
 
 | | Crystal structure of the inward H(+) pump xenorhodopsin | | Descriptor: | Bacteriorhodopsin, EICOSANE, RETINAL, ... | | Authors: | Kovalev, K, Shevchenko, V, Polovinkin, V, Mager, T, Gushchin, I, Melnikov, I, Borshchevskiy, V, Popov, A, Alekseev, A, Gordeliy, V. | | Deposit date: | 2017-11-13 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inward H(+) pump xenorhodopsin: Mechanism and alternative optogenetic approach.
Sci Adv, 3, 2017
|
|
4Y6N
 
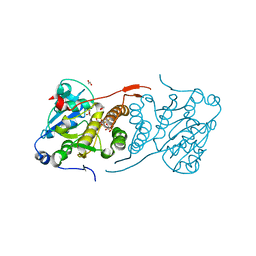 | | Crystal structure of glucosyl-3-phosphoglycerate synthase from Mycobacterium tuberculosis in complex with Mn2+, uridine-diphosphate-glucose (UDP-Glc) and phosphoglyceric acid (PGA) - GpgS Mn2+ UDP-Glc PGA-1 | | Descriptor: | 1,2-ETHANEDIOL, 3-PHOSPHOGLYCERIC ACID, Glucosyl-3-phosphoglycerate synthase, ... | | Authors: | Albesa-Jove, D, Rodrigo-Unzueta, A, Cifuente, J.O, Urresti, S, Comino, N, Sancho-Vaello, E, Guerin, M.E. | | Deposit date: | 2015-02-13 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.348 Å) | | Cite: | A Native Ternary Complex Trapped in a Crystal Reveals the Catalytic Mechanism of a Retaining Glycosyltransferase.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6HP0
 
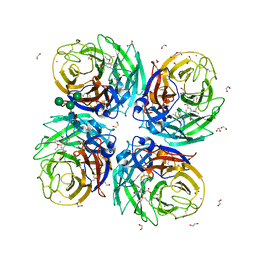 | | Complex of Neuraminidase from H1N1 Influenza Virus in Complex with Oseltamivir Triazol Derivative | | Descriptor: | (3~{R},4~{R},5~{S})-4-acetamido-5-[4-(hydroxymethyl)-1,2,3-triazol-1-yl]-3-pentan-3-yloxy-cyclohexene-1-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pachl, P, Pokorna, J. | | Deposit date: | 2018-09-19 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Investigation of flexibility of neuraminidase 150-loop using tamiflu derivatives in influenza A viruses H1N1 and H5N1.
Bioorg.Med.Chem., 27, 2019
|
|
6VQN
 
 | | Co-crystal structure of human PD-L1 complexed with Compound A | | Descriptor: | N,N'-(2,2'-dimethyl[1,1'-biphenyl]-3,3'-diyl)bis(5-{[(2-hydroxyethyl)amino]methyl}pyridine-2-carboxamide), Programmed cell death 1 ligand 1 | | Authors: | White, A, Lakshminarasimhan, D, Leo, C, Suto, R.K. | | Deposit date: | 2020-02-05 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Checkpoint inhibition through small molecule-induced internalization of programmed death-ligand 1.
Nat Commun, 12, 2021
|
|
6HBS
 
 | |
2J3H
 
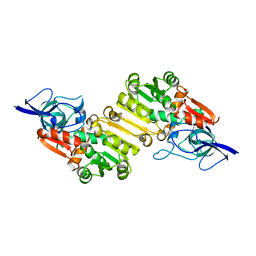 | | Crystal structure of Arabidopsis thaliana Double Bond Reductase (AT5G16970)-Apo form | | Descriptor: | NADP-DEPENDENT OXIDOREDUCTASE P1 | | Authors: | Youn, B, Kim, S.J, Moinuddin, S.G, Lee, C, Bedgar, D.L, Harper, A.R, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2006-08-21 | | Release date: | 2006-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic and Structural Studies of Apoform, Binary, and Ternary Complexes of the Arabidopsis Alkenal Double Bond Reductase at5G16970.
J.Biol.Chem., 281, 2006
|
|
6HBV
 
 | |
