5U6H
 
 | | Solution structure of the zinc fingers 1 and 2 of MBNL1 | | Descriptor: | Muscleblind-like protein 1, ZINC ION | | Authors: | Phukan, P.D, Park, S, Martinez-Yamout, M.M, Zeeb, M, Dyson, H.J, Wright, P.E. | | Deposit date: | 2016-12-08 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Interaction of the Tandem Zinc Finger Domains of Human Muscleblind with Cognate RNA from Human Cardiac Troponin T.
Biochemistry, 56, 2017
|
|
6H53
 
 | |
2N02
 
 | | Solution structure of the A147T variant of the mitochondrial translocator protein (tspo) in complex with pk11195 | | Descriptor: | N-[(2R)-butan-2-yl]-1-(2-chlorophenyl)-N-methylisoquinoline-3-carboxamide, Translocator protein | | Authors: | Jaremko, M, Jaremko, L, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2015-03-04 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Integrity of the A147T Polymorph of Mammalian TSPO.
Chembiochem, 16, 2015
|
|
6B6U
 
 | | Pyruvate Kinase M2 mutant - S437Y | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Srivastava, D, Dey, M. | | Deposit date: | 2017-10-03 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Investigation of a Dimeric Variant of Pyruvate Kinase Muscle Isoform 2.
Biochemistry, 56, 2017
|
|
3A3K
 
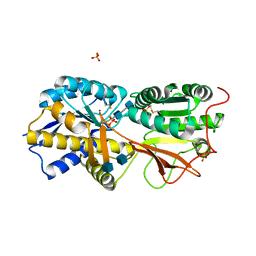 | | Reversibly bound chloride in the atrial natriuretic peptide receptor hormone-binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Atrial natriuretic peptide receptor A, ... | | Authors: | Ogawa, H, Qiu, Y, Ogata, C.M, Misono, K.S. | | Deposit date: | 2009-06-14 | | Release date: | 2010-03-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Reversibly bound chloride in the atrial natriuretic peptide receptor hormone-binding domain: Possible allosteric regulation and a conserved structural motif for the chloride-binding site.
Protein Sci., 19, 2010
|
|
7LXI
 
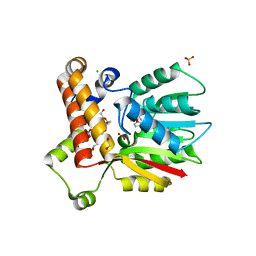 | |
2ZZ9
 
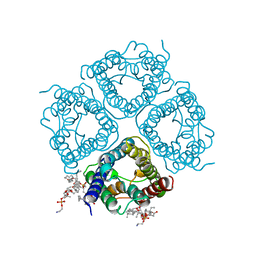 | | Structure of aquaporin-4 S180D mutant at 2.8 A resolution by electron crystallography | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Aquaporin-4 | | Authors: | Tani, K, Mitsuma, T, Hiroaki, Y, Kamegawa, A, Nishikawa, K, Tanimura, Y, Fujiyoshi, Y. | | Deposit date: | 2009-02-06 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-08 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.8 Å) | | Cite: | Mechanism of Aquaporin-4's Fast and Highly Selective Water Conduction and Proton Exclusion.
J.Mol.Biol., 389, 2009
|
|
6B8K
 
 | |
2N0F
 
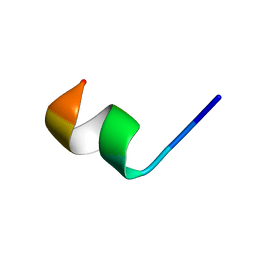 | | NMR structure of Neuromedin C in 60% TFE | | Descriptor: | Neuromedin C (NMC) | | Authors: | Adrover, M, Sanchis, P, Vilanova, B, Pauwels, K, Martorell, G, Perez, J. | | Deposit date: | 2015-03-05 | | Release date: | 2015-10-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Conformational ensembles of neuromedin C reveal a progressive coil-helix transition within a binding-induced folding mechanism.
RSC ADV, 5, 2015
|
|
7LJE
 
 | | Discovery of Spirohydantoins as Selective, Orally Bioavailable Inhibitors of p300/CBP Histone Acetyltransferases | | Descriptor: | 2-[4-[(3'R,4S)-3'-fluoro-1-[2-[(4-fluorophenyl)methyl-[(1S)-2,2,2-trifluoro-1-methyl-ethyl]amino]-2-oxo-ethyl]-2,5-dioxo-spiro[imidazolidine-4,1'-indane]-5'-yl]pyrazol-1-yl]-N-methyl-acetamide, Histone acetyltransferase p300 | | Authors: | Jakob, C.G. | | Deposit date: | 2021-01-29 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.607 Å) | | Cite: | Discovery of spirohydantoins as selective, orally bioavailable inhibitors of p300/CBP histone acetyltransferases.
Bioorg.Med.Chem.Lett., 39, 2021
|
|
5TUS
 
 | | Potent competitive inhibition of human ribonucleotide reductase by a novel non-nucleoside small molecule | | Descriptor: | 2-hydroxy-N'-[(Z)-(2-hydroxynaphthalen-1-yl)methylidene]benzohydrazide, MAGNESIUM ION, Ribonucleoside-diphosphate reductase large subunit, ... | | Authors: | Mohammed, F.A, Alam, I, Dealwis, C.G. | | Deposit date: | 2016-11-07 | | Release date: | 2017-08-02 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Potent competitive inhibition of human ribonucleotide reductase by a nonnucleoside small molecule.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6B7F
 
 | | Crystal structure of E.coli Phosphopantetheine Adenylyltransferase (PPAT/CoaD) in complex with (R)-3,3-dimethyl-4-(5-vinyl-1H-imidazol-1-yl)isochroman-1-one | | Descriptor: | (4R)-4-(5-ethenyl-1H-imidazol-1-yl)-3,3-dimethyl-3,4-dihydro-1H-2-benzopyran-1-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Proudfoot, A.W, Bussiere, D, Lingel, A. | | Deposit date: | 2017-10-03 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.562 Å) | | Cite: | High-Confidence Protein-Ligand Complex Modeling by NMR-Guided Docking Enables Early Hit Optimization.
J. Am. Chem. Soc., 139, 2017
|
|
5TXM
 
 | | STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) TERNARY COMPLEX WITH A DOUBLE STRANDED DNA AND AN INCOMING DDATP | | Descriptor: | 1,2-ETHANEDIOL, 2',3'-dideoxyadenosine triphosphate, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*G)-3'), ... | | Authors: | Das, K, Martinez, S.M, Arnold, E. | | Deposit date: | 2016-11-17 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insights into HIV Reverse Transcriptase Mutations Q151M and Q151M Complex That Confer Multinucleoside Drug Resistance.
Antimicrob. Agents Chemother., 61, 2017
|
|
7M17
 
 | | SN-407-LRRC8A in MSP1E3D1 lipid nanodiscs (Pose-1) | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 7-{[(2S)-2-butyl-6,7-dichloro-2-cyclopentyl-1-oxo-2,3-dihydro-1H-inden-5-yl]oxy}heptanoic acid, Volume-regulated anion channel subunit LRRC8A | | Authors: | Kern, D.M, Gerber, E.E, Brohawn, S.G. | | Deposit date: | 2021-03-12 | | Release date: | 2021-03-24 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Small molecule SWELL1 complex induction improves glycemic control and nonalcoholic fatty liver disease in murine Type 2 diabetes.
Nat Commun, 13, 2022
|
|
6PIS
 
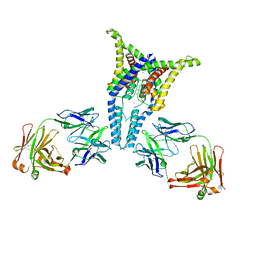 | |
6PJ7
 
 | |
5TWV
 
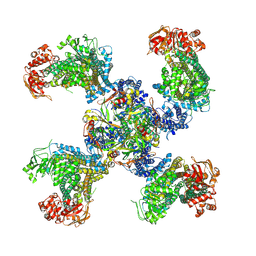 | | Cryo-EM structure of the pancreatic ATP-sensitive K+ channel SUR1/Kir6.2 in the presence of ATP and glibenclamide | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ATP-sensitive inward rectifier potassium channel 11 | | Authors: | Martin, G.M, Yoshioka, C, Chen, J.Z, Shyng, S.L. | | Deposit date: | 2016-11-14 | | Release date: | 2017-01-25 | | Last modified: | 2019-12-25 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Cryo-EM structure of the ATP-sensitive potassium channel illuminates mechanisms of assembly and gating.
Elife, 6, 2017
|
|
2MP8
 
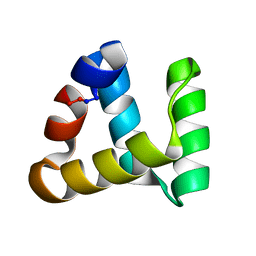 | | NMR structure of NKR-5-3B | | Descriptor: | NKR-5-3B | | Authors: | Rosengren, K.J, Craik, D.J. | | Deposit date: | 2014-05-13 | | Release date: | 2015-05-13 | | Last modified: | 2016-06-01 | | Method: | SOLUTION NMR | | Cite: | Identification, Characterization, and Three-Dimensional Structure of the Novel Circular Bacteriocin, Enterocin NKR-5-3B, from Enterococcus faecium
Biochemistry, 54, 2015
|
|
6PP2
 
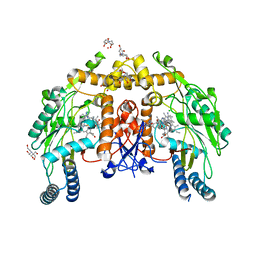 | | Structure of human endothelial nitric oxide synthase heme domain in complex with 4-((4-(2-Amino-4-methylquinolin-7-yl)-2-(aminomethyl)phenoxy)methyl)benzonitrile | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-{[2-(aminomethyl)-4-(2-amino-4-methylquinolin-7-yl)phenoxy]methyl}benzonitrile, 5,6,7,8-TETRAHYDROBIOPTERIN, ... | | Authors: | Chreifi, G, Li, H, Poulos, T.L. | | Deposit date: | 2019-07-05 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | First Contact: 7-Phenyl-2-Aminoquinolines, Potent and Selective Neuronal Nitric Oxide Synthase Inhibitors That Target an Isoform-Specific Aspartate.
J.Med.Chem., 63, 2020
|
|
7JLU
 
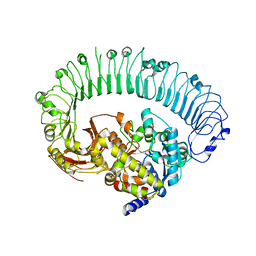 | | Structure of the activated Roq1 resistosome directly recognizing the pathogen effector XopQ | | Descriptor: | CALCIUM ION, Disease resistance protein Roq1, XopQ | | Authors: | Martin, R, Qi, T, Zhang, H, Lui, F, King, M, Toth, C, Nogales, E, Staskawicz, B.J. | | Deposit date: | 2020-07-30 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the activated ROQ1 resistosome directly recognizing the pathogen effector XopQ.
Science, 370, 2020
|
|
5TZR
 
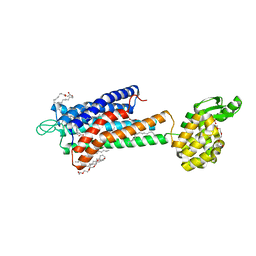 | | GPR40 in complex with partial agonist MK-8666 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (5aR,6S,6aS)-3-({2',6'-dimethyl-4'-[3-(methylsulfonyl)propoxy][1,1'-biphenyl]-3-yl}methoxy)-5,5a,6,6a-tetrahydrocyclopropa[4,5]cyclopenta[1,2-c]pyridine-6-carboxylic acid, Free fatty acid receptor 1,Endolysin,Free fatty acid receptor 1, ... | | Authors: | Lu, J, Byrne, N, Patel, S, Sharma, S, Soisson, S.M. | | Deposit date: | 2016-11-22 | | Release date: | 2017-06-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the cooperative allosteric activation of the free fatty acid receptor GPR40.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5U0B
 
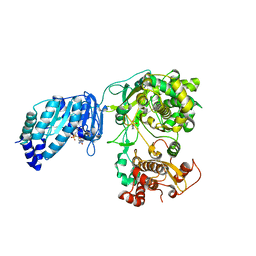 | | Structure of full-length Zika virus NS5 | | Descriptor: | Genome polyprotein, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Zhao, B, Du, F. | | Deposit date: | 2016-11-23 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and function of the Zika virus full-length NS5 protein.
Nat Commun, 8, 2017
|
|
6PN4
 
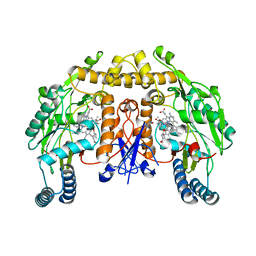 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 7-(3-(Aminomethyl)-4-(cyclopropylmethoxy)phenyl)-4-methylquinolin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[3-(aminomethyl)-4-(cyclopropylmethoxy)phenyl]-4-methylquinolin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2019-07-02 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | First Contact: 7-Phenyl-2-Aminoquinolines, Potent and Selective Neuronal Nitric Oxide Synthase Inhibitors That Target an Isoform-Specific Aspartate.
J.Med.Chem., 63, 2020
|
|
2N0G
 
 | | NMR structure of Neuromedin C in 90% TFE | | Descriptor: | Neuromedin C (NMC) | | Authors: | Adrover, M, Sanchis, P, Vilanova, B, Pauwels, K, Martorell, G, Perez, J. | | Deposit date: | 2015-03-05 | | Release date: | 2015-10-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Conformational ensembles of neuromedin C reveal a progressive coil-helix transition within a binding-induced folding mechanism.
RSC ADV, 5, 2015
|
|
6BBH
 
 | |
